Dania Al Ismail1,*, Edgar I. Campos-Madueno1,*, Valentina Donà2, Andrea Endimiani1
1Institute for Infectious Diseases (IFIK), University of Bern, Bern, Switzerland
2Independent Researcher and Scientific Writer, Bolzano, Italy
*Authors contributed equally
Prof. Andrea Endimiani, MD, PhD, FAMH, FISAC
andrea.endimiani@unibe.ch
Al Ismail D, Campos-Madueno EI, Donà V, Endimiani A. Hypervirulent Klebsiella pneumoniae (hvKp): Overview, Epidemiology, and Laboratory Detection. Pathogens and Immunity. 2025;10(1):80–119. doi: 10.20411/pai.v10i1.777
10.20411/pai.v10i1.777
Klebsiella pneumoniae (Kp) is a Gram-negative pathogen responsible for both hospital- and community-acquired infections. Kp is classified into 2 distinct pathotypes: classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp). First described in Taiwan in 1986, hvKp are highly pathogenic and characterized by unique phenotypic and genotypic traits. The hypermucoviscous (hmv) phenotype, generally marked by overproduction of the capsule, is often associated with hvKp, although recent studies show that some cKp strains may also have this characteristic. Furthermore, hvKp can cause severe community-acquired infections in healthy people and have been associated with metastatic infections such as liver abscess, meningitis, and endophthalmitis. HvKp are increasingly being reported in hospital-acquired settings, complicating treatment strategies. In particular, while hvKp have historically been antibiotic-susceptible, multidrug-resistant (MDR) strains have emerged and pose a significant public health threat. The combination of high virulence and limited antibiotic options demands further research into virulence mechanisms and rapid identification methods.
This review discusses the epidemiology of hvKp and their virulence factors, highlighting the importance of phenotypic and non-phenotypic tests, including next-generation molecular diagnostics, for the early detection of hvKp.
Klebsiella pneumoniae, virulence, hvKp, epidemiology, detection, ESBL, carbapenemases, string test, hypermucoviscous, WGS, in vivo
Klebsiella pneumoniae (Kp) is a well-known Gram-negative opportunistic pathogen within the Enterobacteriaceae family that is frequently responsible for both hospital- and community-acquired infections [1–3]. Kp is classified into 2 distinct pathotypes – classical K. pneumoniae (cKp) and hypervirulent K. pneumoniae (hvKp) – which vary in their clinical presentation, epidemiology, identification methods, and management strategies (Table 1) [4–6].
Since its first description in 1986 in Taiwan [7], hvKp has emerged worldwide as a particularly virulent pathotype with unique phenotypic and genotypic characteristics [8–10]. For instance, a trait that is usually associated with hvKp is a hypermucoviscous (hmv) phenotype with an overproduction of the capsule [11–13]. However, recent studies have found that not all hvKp strains are hmv and some cKp strains may also possess this characteristic [4, 5, 14, 15].
While cKp is a frequent cause of primary pneumonia and urinary tract infections in the hospital setting (especially in the elderly or immunocompromised patients [16, 17]), hvKp is more virulent and can cause community-acquired infections in healthy individuals, even though hospital-acquired infections are being increasingly reported as well [4, 18]. Besides its role as a main causative agent of pyogenic liver abscess [19], hvKp may also lead to other unusual and multiple body-site infections (ie, metastatic spread), such as endophthalmitis, meningitis, septic arthritis, lung abscess, pneumonia, epidural abscess, osteomyelitis, non-hepatic abscesses, and necrotizing fasciitis [9, 11, 15, 18, 20–24]. Overall, infections due to hvKp have high rates of morbidity and mortality [25–27]. Therefore, prompt therapeutic interventions are essential to prevent a poor prognosis [2, 15, 28].
The prevalence of the hypervirulent pathotype amongst Kp is quite variable, but it may be as high as 12% to 45% in endemic areas like China [29–31]. Complicating matters is the fact that, in the past, hvKp isolates were primarily susceptible to antibiotics, but many recent studies report the emergence of multidrug-resistant (MDR) strains [32]. As a consequence, the combination of high virulence and limited antibiotic treatment options represents an ultimate threat to our public health systems. This concerning fact has spurred increased research attention towards this pathogen, focusing on identifying its main virulence markers. Therefore, understanding the virulence mechanisms underlying this pathotype and its rapid identification are crucial for the development of effective treatments and preventive measures.
This review provides an overview of the epidemiology of hvKp and underscores the importance of investigating their virulence factors (VFs). Furthermore, we explore laboratory detection methods – with particular emphasis on molecular diagnostics – to facilitate rapid identification of infections caused by hvKp strains.
Table 1. General Characteristics of Hypervirulent and Classical Klebsiella pneumoniae Strains
|
Features |
hvKp |
cKp |
|
Acquisition of infections |
Community |
Hospitals and long-term care facilities |
|
Population affected |
All ages, healthy individuals |
Elderly, immuno- compromised |
|
Number of infection sites |
Multiple |
Single |
|
Metastasis or additional site |
Common |
Uncommon |
|
Liver abscess |
Not associated with biliary disease |
Associated with biliary disease |
|
Unusual infectious |
Endophthalmitis, meningitis, septic arthritis, splenic abscess, necrotizing fasciitis |
Rare |
|
Hmv phenotype |
Usual |
Rare |
|
Geographic spread |
Worldwide |
Worldwide |
|
Serotypes identified |
K1, K2, K5, K16, K20, K54, K57, KN1 |
K1 to K79 |
|
Siderophores production |
Aerobactin, salmochelin, enterobactin, yersiniabactin |
Enterobactin, yersiniabactin |
|
Acquired virulence-encoding genes |
magA (mucoviscosity-associated gene A), rmpA, rmpA2 (regulators for capsule), peg-344 (inner membrane transporter), iucA, iutA (aerobactin biosynthesis), iroB (salmochelin biosynthesis), clb (colibactin), terW (tellurite resistance) |
May possess some of them [33] |
Note. hvKp, hypervirulent K. pneumoniae; cKp, classical K. pneumoniae; hmv, hypermucoviscous; ICE, integrative conjugative element
Factors that may contribute to Kp hypervirulence, such as capsular hyperproduction, hmv phenotype, siderophores, and other potential contributors (eg, lipopolysaccharides (LPS), colibactin and fimbriae) have been extensively studied since its first emergence 4 decades ago. So far, several biomarkers associated with hypervirulence have been identified more frequently in hvKp than cKp strains (Table 1), including genetic components on chromosomes, virulence plasmids, or a combination of both [27, 34]. Of note, most of the main VFs of hvKp are usually located on plasmids or within mobile genetic elements (MGEs) integrated in the chromosome, suggesting that horizontal gene transfer (HGT) may be an important mechanism for the development of a hypervirulent phenotype [35].
The production of peculiar capsular polysaccharides is a crucial factor for Kp survival within the host and represents a key virulence determinant for its capacity to evade phagocytosis, complement, antimicrobial peptides, and specific antibodies [2, 18, 36–38]. In this context, the serological classification of Kp typically relies on capsule (ie, K antigen) serotyping, with at least 79 distinct capsule types identified to date [33, 39–41]. In particular, 8 types (K1, K2, K5, K16, K20, K54, K57, and KN1) have been described in hvKp (Table 1) [36, 42–44], with K1 and K2 being the most frequently reported [18, 27, 36, 45].
Basic capsule production in Kp is a process regulated by genes located in the capsule polysaccharide synthesis (cps) locus of the chromosome, including genes such as wzi, wza, wzb, wzc, gnd, wca, cpsB, cpsG, and galF [46].
Capsule overproduction is influenced by several chromosomal genes, such as the mucoviscosity-associated gene A (magA; newly termed as wzy-K1, ie, the serotype K1 polymerase gene in the cps locus) and both crmpA and crmpA2 genes (encoding regulators of the mucoid phenotype). Moreover, capsule overproduction can also be influenced by the plasmid-mediated genes prmpA, prmpA2, and peg-344 (encoding a putative transporter; see below) that can be found with different frequencies on virulence plasmids (eg, pLVPK with all 3 genes; pVir-CR-hvKp4 with only prmpA2) [11, 46]. Interestingly, rmpA and peg-344 can also be found in the integrative conjugative element (ICE) of serotype K1 Kp (ICEKp1). Overall, the above chromosomal or plasmidic genes are regarded as valid molecular markers for the detection of hvKp, although cKp may also rarely possess some of them (Table 1) [15, 22, 27, 33, 36, 47–49]. Particularly, rmpA is considered specific for hvKp identification due to its significant contribution in hypercapsule production, enhancing the pathogenicity of hvKp [11, 48].
The most striking feature of the majority of hvKp is the hypermucoviscous (hmv) phenotype, a trait that is usually associated with excess capsule synthesis (see above). However, the real link between hmv phenotype and capsule overproduction is still under investigation.
Indeed, the hmv phenotype is not solely determined by capsular polysaccharide overproduction, but is also associated with additional factors, such as the presence of rmpC and rmpD genes (ie, capsular and hmv regulators, respectively) located on the virulence plasmids [50, 51]. In this context, a ΔrmpC Kp mutant was shown to retain hypermucoviscosity but produced less capsule compared to the parental strain. In contrast, the ΔrmpD mutant exhibited no difference in capsule production but lost its hmv phenotype. These results are consistent with some early observations suggesting that capsule overproduction and hypermucoviscosity are probably regulated in separate manners [52].
Of note, not all hvKp isolates display the hmv phenotype, which can result in a negative string test (see below). For instance, a study conducted in China involving 47 Kp strains associated with liver abscesses (usually due to hvKp) showed by PCR analyses that all strains carried the virulence genes rmpA/rmpA2, as well as other virulence genes linked to hypervirulence, such as iucA and iroB encoding the siderophores aerobactin and salmochelin, respectively (see below). Based on these results, all strains were defined as hvKp, although only 31.9% (15/47) of them exhibited the hmv phenotype by using the string test [53].
Siderophores are small low-molecular-weight chelators that play a crucial role in bacterial Fe acquisition. These molecules are secreted by the bacteria into the external environment, where they bind Fe with extremely high affinity and are then taken back into the bacterial cells via specific receptors and transport systems [36]. This process provides the bacteria with the Fe they need to grow and is considered an important VF [18], as it allows the bacteria to survive in the usually Fe-poor environment of the infection site resulting from a process known as nutritional immunity (ie, a series of different Fe-limiting strategies used by the host immune system to protect itself) [54].
Indeed, Kp strains with an in vitro siderophore production greater than 30 μg/mL have been linked to severe illness and mortality in a mouse systemic infection model [11]. Therefore, this quantitative difference in siderophore production may aid in the discrimination between hvKp and cKp strains, with hvKp exhibiting higher siderophore production levels (see below) [47, 55–58].
Kp strains may produce 4 different types of siderophores: aerobactin, salmochelin, yersiniabactin, and enterobactin. Their expression level is linked to the iuc, iro, ybt, and ent gene clusters, respectively [11, 18, 59]. In particular, targeting aerobactin and salmochelin is of great importance to recognize hvKp strains.
Aerobactin is expressed in over 90% of hvKp strains and is more specific for this pathotype compared to enterobactin and yersiniabactin, which are often found in both cKp and hvKp [6]. This underscores the critical role of these siderophores as an essential VF in hvKp, particularly in systemic infections [60]. Of note, the genes responsible for aerobactin biosynthesis and transport (iucABCD and iutA operon) are located on hvKp virulence plasmids (eg, pLVPK and pVir-CR-hvKp4 carry both genes) [36, 61, 62]. Salmochelin, expressed by genes of the iroA locus (eg, iroBCDN) is also often found and highly expressed in hvKp. Therefore, iroA is considered another main VF for hvKp strains [36, 41, 46]. Interestingly, a recent analysis of ~2,500 genomes suggested a high co-occurrence of the iuc and iro loci in hvKp [63].
In a study by Sheng et al, all Kp causing bacteremia and testing positive for the aerobactin encoding gene resulted to be hvKp strains, and well above half of such isolates carried genes coding for salmochelin (86.2%) and yersiniabactin (72.4%) [59]. In another study considering 97 Kp genomes of the hypervirulent sequence type (ST) 23, genes encoding all 3 siderophores were found in almost all genomes [64].
In addition to the previously discussed VFs, hvKp may possess further virulence-linked traits such as LPS, colibactin, fimbriae, and peg-344.
LPS is composed of lipid A, an oligosaccharide core, and the O antigen. These components, altogether also known as endotoxin, are encoded by lpx, waa, and wb gene clusters, respectively, in all Kp strains [36, 46]. LPS functions as a protective barrier against humoral defenses even in the presence of the capsule and also acts as a potent immune activator [11]. Currently, the specific role of LPS produced by hvKp strains in their enhanced virulence compared to cKp remains uncertain [41].
Colibactin is a genotoxic metabolite expressed by genes (clb) located on the polyketide synthase genomic island (pks), typically found within a chromosomal ICE [18]. The presence of the pks island is well-documented in certain strains of Escherichia coli, and it has been increasingly recognized in Kp, including hvKp. Colibactin induces DNA damage in host cells and appears to contribute to the colonization and pathogenesis of hvKp [11, 27, 46, 59].
Analysis of Kp fimbriae has identified two key types: type 1 and type 3 [36]. These fimbriae are encoded by the fimH (type 1) and the mrkABCD (type 3) gene clusters, and both play crucial roles in biofilm formation [46]. In particular, type 3 fimbriae are recognized as a significant bacterial VF in hvKp, however, their role in hvKp remains poorly understood. Alharbi et al showed that biofilm formation in hvKp isolates was significantly higher than in cKp strains [65, 66]. In contrast, other studies found that hvKp strains were associated with weak biofilm formation [67, 68], showing no significant difference from cKp in this regard [69].
As anticipated above, peg-344 can be carried by some MGEs in hvKp strains (eg, plasmid pLVPK, but not pVir-CR-hvKp4). It encodes a metabolic transporter with an unknown function located within the inner membrane. When Kp is grown in human ascites, the expression of peg-344 is increased [11, 46]. Additionally, peg-344 was essential for full virulence in a murine model of pneumonia challenge but did not appear to be related to the induction of sepsis after subcutaneous challenge [70].
Several authors reported rare hvKp strains lacking the common regulators of the hmv phenotype (rmpA/rmpA2) or the siderophores [71, 72][73]. For instance, a ST14 MDR Kp isolate in Sudan did not possess rmpA and rmpA2, although it showed an hmv phenotype based on the positive string test. The strain was also of serotype K2 and produced aerobactin and salmochelin, but the hypervirulent phenotype was not analyzed and confirmed with an in vivo model [72]. Similarly, 2 MDR-Kp strains (ST15 and ST307) from China were reported as hmv and hypervirulent by using the string test or the mucoviscosity assay and the Galleria mellonella infection model, respectively. Notably, both strains lacked rmpA, rmpA2, aerobactin, and salmochelin genes [73]. Overall, it seems that the hmv/hypervirulent phenotypes of hvKp can also be determined by non-classic biomarkers that should be further investigated with appropriate methodologies. In this context, we emphasize that the implementation of the string test or the G. mellonella in vivo model may not be appropriate to define the hmv/hypervirulent phenotypes (see below).
Historically, hvKp strains were typically sensitive to common antibiotic agents, including last-generation cephalosporins and carbapenems. However, in the last years, several concerning reports of the emergence of MDR hvKp strains acquiring different ARGs through HGT of MGEs have been published [32, 35, 64, 74]. In particular, cases of hvKp producing extended-spectrum β-lactamases (ESBLs) and/or carbapenemases have been documented [3, 75].
In this context, there are different mechanisms that can mediate this phenomenon [11, 76]: 1) ESBL- and/or carbapenemase-producing (CP) cKp strains acquire a virulence plasmid (eg, pLVPK) which is typically non-conjugative but can be acquired through co-mobilization by helper conjugative plasmids (usually of IncF type); 2) hvKp strains acquire a conjugative plasmid that carries ESBL and/or carbapenemase encoding bla genes; 3) cKp strains acquire both ESBL and/
or carbapenemase encoding bla genes along with the VFs via a virulence hybrid plasmid resulting from the recombination between a resistance and a virulence plasmid [11, 77–83].
Most recent studies focus on the detection and description of CP hvKp strains (see below), while only few performed in the past provide data regarding ESBL-producing hvKp strains. In a survey conducted in China (2013), 37% (85/230) of the Kp strains were identified as hvKp based on the presence of rmpA. Among them, 13% (11/85) were found to be ESBL producers, with CTX-Ms being the most frequently detected ESBLs [84]. Similarly, Liu et al indicated that 45.7% (80/175) of strains collected in China between 2008 and 2014 were predicted to be hvKp based on the presence of K1 and K2 capsule types or magA, rmpA, and rmpA2 genes. Among them, 16.3% (13/80) were ESBL producers [85].
The clinical impact of ESBL-producing hvKp strains is significant, as evidenced by a series of cases involving liver abscess, meningitis, and severe infections [86, 87]. For instance, Xu et al described the first case of endophthalmitis in China associated with a CTX-M-14-producing hvKp strain belonging to clonal complex (CC) 23 and K1 serotype; the blaCTX-M-14 gene was acquired through horizontal transmission of a plasmid. The patient’s visual activity recovered only after prolonged imipenem treatment [88].
Carbapenem resistance in Kp involves various mechanisms, with carbapenemase production being the most significant because of its association to MGEs [76, 89]. This phenomenon is also true for the carbapenem-resistant (CR) hvKp strains [90].
The first CP hvKp strain (belonging to K2 and ST65) was identified in China in 2015 due to the acquisition of a plasmid harboring a blaKPC carbapenemase gene [91]. To date, the emergence of CP hvKp strains has been documented worldwide. In this context, the acquisition of a KPC encoding gene is the most common [44, 46, 92–95], but NDM- [29, 96, 97], and OXA-48-like CP strains have also been reported [80, 98].
For instance, Yang et al conducted a screening of 784 KPC-2-producing Kp strains in 3 hospitals between 2014 and 2017 in China. They investigated the presence of virulence genes (ie, rmpA, rmpA2, and iutA) revealing that 457 (58%) strains harbored at least 2 of them [99]. In another study from China, 69 non-duplicated CR Kp isolates were collected during 2021 to 2022 and analyzed by PCR. As a result, 27 out of 69 (39.1%) strains were considered hvKp based on the presence of rmpA and/or rmpA2 with iucA, iroB, and peg-344. Among these, 22 (81.5%) carried blaKPC-2 and belonged to ST11 [100]. In a study from Germany with CP Kp strains collected between 2013 and 2021, the iuc gene (aerobactin) was present in ١٨ of 109 (16.5%). These isolates produced OXA-232 (n=7), OXA-48 (n=6), OXA-48 and NDM (n=3), NDM (n=1), and KPC (n=1) carbapenemases. Seven out of 18 CP hvKp isolates were also resistant to ceftazidime-avibactam, colistin, and/or cefiderocol [80]. In this context, we emphasize that hvKp strains producing KPC variants (eg, KPC-31, KPC-135) conferring resistance to ceftazidime-avibactam are emerging [101, 102].
Many studies have identified hvKp strains in clinical samples from humans and animals, but these pathogens are nowadays also reported in contaminated food and the environment [41, 103]. Historically, these studies predominantly originated from Southeastern Asia; however, reports from other areas, including Europe and the United States started to become more frequent in the last 10 years [104–106].
HvKp strains, particularly MDR hvKp, can cause severe infections and lead to outbreaks with high mortality rates, particularly in immunocompetent individuals [27]. For instance, hvKp bacteremia has a mortality rate up to 37% [107]. In addition, patients with metastatic disease may encounter serious long-term neurological or visual disabilities [108, 109].
Most hvKp strains belong to a relatively small group of clones, indicating the importance of a particular genetic background for the development of hypervirulence. For instance, many studies have shown that strains belonging to the CC23 (particularly the ST23 of capsular serotype K1) and the ST65 and ST86 (both of capsular serotype K2) are frequently associated with hvKp [26, 103, 110]. MDR hvKp strains (especially those producing carbapenemases) are rather linked to other STs, like ST11 (K24/K47/K64), ST15 (K24/K47/K54/K64/K112), ST101 (K62), ST147 (K20/K64), ST231 (K51), ST258 (K106/K107), ST307 (K102), ST395 (K2/K39), and ST512 (K107) [80, 111, 112].
Infections with hvKp are geographically widespread. Initially, hvKp infections were solely documented in the Asia-Pacific region, including China and Singapore [113, 114], with prevalence rates between 12% to 45% during 2008 to 2017 [29–31]. However, at the beginning of the 21st century, there has been a global expansion of hvKp cases, reaching Europe and North America. For example, a study conducted in Spain (2007-2013) found that 53 out of 878 (5.4%) isolates exhibited an hmv phenotype by the string test and belonged to the K1/K2 serotypes, of which 16 (1.8%) being ST23/K1 [26]. Similar trends have been observed in the United States (6.3% in 2009-2010) and Canada (8.2% in 2001-2007) [115, 116]. According to the European Centre for Disease Prevention and Control, most of the contemporary hvKp reported from healthcare facilities in the European Union/European Economic Area countries belong to the ST23/K1 clone [117].
With such an increasing incidence of hvKp infections, it is also necessary to focus on the intestinal colonization of hvKp, which can directly precede subsequent infections in healthy people. Numerous studies have confirmed that the human gut is a reservoir for Kp [118–120]. However, accurate assessment of hvKp colonization rates in the community is challenging due to the inconsistent use of hvKp-specific markers to distinguish them from cKp. Similar to cKp, colonization by hvKp is likely, but it does not always lead to subsequent infection [11].
Yang et al analyzed data from public databases, revealing that hvKp can indeed colonize the gut of Chinese individuals. They collected 95 gut Kp isolates from 69 healthy individuals and found that the carriage rate of hvKp among these individuals was 12% as evidenced by the presence of the iuc locus and rmpA/rmpA2 genes detected by PCR [121]. Observations of hvKp gut colonization have also been reported in other countries. A study among healthy Koreans revealed a hvKp colonization rate of 4.6% in 2007 [122]. In Norway (2015-2016), 5 out of 484 (1%) Kp strains isolated from fecal samples were defined as hvKp [123]. Between 2004 and 2010, in Malaysia, Singapore, Taiwan, Japan, and Thailand, colonization rates were 14.1%, 14.9%, 11.3%, 16.7%, and 2.7%, respectively [11, 47].
In a recent report from Egypt, the overall prevalence of hvKp and CR hvKp in diarrheic farm animals was 7.9% and 6.1%, respectively. Of note, hvKp and CR hvKp were detected among all examined farm animal species (cattle, sheep, and goats) [124]. In another study performed in China (2017-2019), the screening of companion animals (dogs and cats) found an overall Kp prevalence of 2.0% (n=105). Moreover, 8.6% of the Kp strains (n=9) had an hmv phenotype, while 64 isolates (61%) were hvKp based on the G. mellonella infection model (see below) [125].
HvKp have been also isolated from different food and environmental sources. In a study from China (2017), 2 ST23 hvKp carrying blaKPC-2 were isolated from cucumbers in ready-to-eat vegetable samples [126]. Li et al (China) reported the presence of the tigecycline resistance gene tet(X4) in hvKp strains from pork samples collected from multiple markets in 2020 [127]. Three MDR hvKp strains were isolated from public water environments in Brazil (2018), highlighting another potential source of hvKp acquisition [128]. In Egypt, CP hvKp strains have been isolated from fresh oysters [129]. These data support the need for a One Health approach, including surveillance of non-human settings, to reduce the spread of hvKp.
For many years, infections due to hvKp have been diagnosed solely on the basis of clinical presentation [11]. Meanwhile, several phenotypic and genotypic diagnostic tests have been developed in an attempt to specifically detect hvKp, which vary widely in methodology and accuracy (Table 2).
We emphasize that a consensus definition of hvKp does not yet exist. This controversial issue affects the interpretation of epidemiological data (see above) and also the comparison of the analytic performance of diagnostic tests described below [56, 106]. In this context, we believe that accurate and powerful studies combining whole-genome sequencing analysis of key biomarkers (eg, iucA, rmpA, rmpA2, iroB, peg-344) and in vivo murine testing (eg, Russo et al [33]) will provide essential insights to elaborate a standardized definition for hvKp.
Mouse killing assay. The mouse lethality assay is an accurate method for identifying hvKp and differentiating it from cKp strains. Despite its accuracy, the assay is time-consuming, costly, and complex, limiting its widespread application (Table 2). However, mouse infection models remain a standard for assessing pathogen virulence, including that for Kp. Typically, the lethal dose (LD) for hvKp in a mouse model is less than 106 colony forming units (CFUs), while for cKp it exceeds 107 CFU [130]. The assay can be performed using either intraperitoneal (IP) or subcutaneous (SC) injection methods.
Table 2. Available Methods for the Assessment of the Pathotype of K. pneumoniae: Main Advantages and Disadvantages
|
Detection tests |
Method |
Advantages |
Disadvantages |
|
In vivo models |
Mouse killing assay |
|
|
|
G. mellonella infection model |
|
|
|
|
Phenotypic tests |
String test |
|
|
|
Siderophore production tests |
|
|
|
|
Mucoviscosity assay |
|
|
|
|
Serum killing assay |
|
|
|
|
Tellurite resistance test |
|
|
|
|
Non-phenotypic tests |
Multiplex qPCR |
|
|
|
LAMP (Eazyplex® hvKp assay) |
|
|
|
|
RAA |
|
|
|
|
MALDI-TOF MS |
|
|
|
|
ICT (ICS) |
|
|
|
|
Raman spectroscopy |
|
|
|
|
NGS Illumina |
|
|
|
|
NGS Oxford Nanopore/PacBio |
|
|
|
|
mNGS |
|
|
Note. Multiplex qPCR, multiplex quantitative polymerase chain reaction; LAMP, loop-mediated isothermal amplification; RAA, recombinase-aided amplification; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; ICT, immunochromatographic test; ICS, immunochromatographic strip; NGS, next-generation sequencing technologies; TAT, turnaround time; mNGS, metagenomics NGS; Kp, K. pneumoniae; hvKp, hypervirulent K. pneumoniae; cKp, classical K. pneumoniae.
In one study, mice were infected with 100 µL of varying concentrations (103 to 106 CFU) from six hvKp strains. Mortality was monitored over 7 days. The 50% lethal dose (LD50) for 3 strains (2 K2 and 1 K1) was found to be less than 102-103 CFU, while the remaining 3 strains (K57, K105, and another K1) had an LD50 of 104-105 CFU. The LD50 for K1 strains was 102-105 CFU, while for K2 strains it was 102-103 CFU [53]. Wang et al conducted a study showing significant variation in LD50 among hvKp strains. They injected mice with 100 µL of bacterial suspension intraperitoneally and observed mortality for 14 days. Mice challenged with K1/K2 isolates showed an LD50 ranging from ≤102 to 2x103 CFU, whereas mice infected with ST11 isolates with serotypes K20, K47, and K64 exhibited no illness symptoms with a high LD50 of >107 CFU [131].
Russo et al found variability in virulence among hvKp strains [57]. After SC challenges of CD1 mice, the strains were categorized based on the lethal challenge inoculum (CI) as follows: fully virulent hvKp (fvhvKp) strains that were lethal at a CI ≤10³ CFU, partially virulent hvKp (pvhvKp) strains that were lethal at a CI between 10⁴ and 10⁷ CFU, and cKp strains that were not lethal even at a CI of 10⁷ CFU. Mortality rates for fvhvKp after SC challenges were 80% at 10² CFU and 100% at 10³ CFU, compared to pvhvKp, which showed 0% mortality at both 10² and 10³ CFU. However, at CIs of 10⁴ to 10⁷ CFU, pvhvKp mortality rates were 20%, 70%, 80%, and 80%, respectively. For IP challenges in CD1 mice, mortality rates for fvhvKp were 40% at 10² CFU and 60% at 10³ CFU. In contrast, challenge with an inoculum of 10⁴ CFU of pvhvKp showed 0% mortality in 2 mouse types (ie, BALB/c and CD1) and 50% mortality in 1 (C57BL/6), indicating that IP challenges of BALB/c and CD1 mice, but not C57BL/6 mice, may be used to clearly differentiate fvhvKp from pvhvKp. In conclusion, both SC and IP challenge models effectively distinguished between hvKp and cKp strains. Of note, no mortality was observed in SC challenges with cKp challenge inoculum up to 108 CFU, while IP challenges with cKp had an LD50 >107 CFU [57].
In another study, mice were injected subcutaneously with concentrations ranging from 2x103 to 5x103 CFU for both hvKp and cKp strains and with CI of 3x107 to 6x107 CFU for cKp strains, monitored over 14 days. The mean 14-day death rates for hvKp and cKp strains were 91.2% and 0% with CI of 2x103 to 5x103 CFU, respectively. The mortality rate for the cKp strain remained at 0% even with 4-log higher CIs of 3x107 to 6x107 CFU [132].
Galleria mellonella infection model. The G. mellonella infection model has been employed to investigate various bacteria and assess the virulence of Kp due to the ease and cost-effectiveness of obtaining larvae (Table 2) [9]. Typically, 10µL to 20µL of a specific concentration of Kp is injected into G. mellonella larvae through the left second hind proleg, which are then maintained at 37°C in darkness. Larval survival is monitored daily, and death is determined when larvae cease responding to mechanical stimuli and exhibit a change in body color from yellowish to black (ie, melanization), with the time of death recorded [9].
Li et al demonstrated that using the G. mellonella infection model alone for identifying hvKp yielded a sensitivity of 97.8% and a negative predictive value (NPV) of 95.2%. However, the specificity and positive predictive value (PPV) were notably lower at 34.5% and 53.6%, respectively. Combining the G. mellonella infection model with the string test led to a significantly improved sensitivity, specificity, PPV, and NPV (95.6%, 94.8%, 93.5%, and 96.5%, respectively) [130].
The use of G. mellonella to differentiate hvKp from cKp was also investigated in other studies. Russo et al found that when a CI of 1x105 CFU was administered, the 5-day mortality rates for hvKp and cKp strains were 93.4% and 71%, respectively. However, for CI ranging from 1x104 to 5x104 CFU and 1x105 to 5x105 CFU, the differences in mortality were minimal (25.8% and 22.4%, respectively). This indicates that the G. mellonella model alone may not be a reliable method for differentiating hvKp from cKp [132].
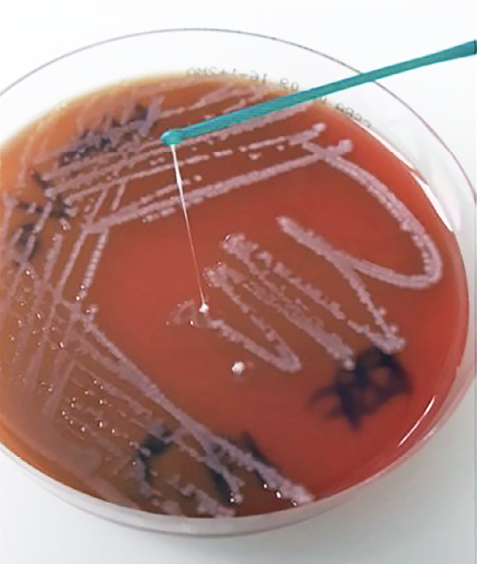
String test. This test is classically used to determine the hmv phenotype of Kp, a trait that typically characterizes most hvKp. This simple test involves touching a bacterial colony on a blood agar plate with an inoculation loop and then pulling the loop away (Figure 1) [1, 9]. A positive result is indicated by the formation of a viscous string stretching from the bacterial colony to the loop measuring ≥5 mm [15, 43]. However, recent research indicates that the string test, which is still widely used as the primary assay to identify hvKp strains, is less effective than other methods.
For instance, Russo et al found that the string test resulted in an accuracy, sensitivity, and specificity of 90%, 89%, and 91%, respectively, which were inferior to the testing values obtained for several other genotypic biomarkers (ie, peg-344, iroB, iucA, rmpA, and rmpA2) [55]. In another study, the positive rate of the string test was only 13.8%, which was equal to or lower than the positive detection rates obtained by PCR analysis individually targeting the 5 major virulence genes peg-344, iroB, iucA, rmpA, rmpA2 (ie, 43.6%, 19.2%, 50%, 10.6%, and 50%, respectively) [133]. In a further study involving 56 strains, previously identified as hvKp based on the clinical presentation (ie, from patients with liver abscesses), only 64.3% (n=36) exhibited a positive hmv phenotype by string test. Among these 36 hmv strains, 100% were positive for the rmpA gene by PCR. However, 95% of the non-hmv hvKp strains were also positive for rmpA, suggesting that this gene may be necessary, but not sufficient for the development of the hmv trait [134].

Figure 1. String test on Columbia agar with 5% sheep blood (CSBA, Oxoid). Since a string with a length >5 mm was observed, the test result was considered positive (ie, hypermucoviscous phenotype of the K. pneumoniae strain).
In conclusion, reliance on the string test alone appears inadequate for the identification of hvKp due to its suboptimal sensitivity and specificity (ie, negative and positive string test results may also be reported for hvKp and cKp strains, respectively) (Table 2) [5, 135–137].
Siderophore tests. Assessing siderophore production shows promise for differentiating hvKp from cKp strains. Both qualitative and quantitative methods could be used to evaluate siderophore production by the bacterial strains (Table 2) [33].
In the qualitative siderophore production test, strains are grown overnight at 37°C in M9 iron-chelated medium containing casamino acids (c-M9-CA). After centrifugation, the supernatants are diluted 5-fold in c-M9-CA. These are then mixed with a 98% chrome azurol S (CAS)-containing siderophore assay solution and incubated in the wells of a flat-bottomed 96-well plate in the dark for 30 minutes. A color change from blue to orange indicates the production of siderophores, signifying a positive test result. Russo et al employed this method to analyze and differentiate between cKp and hvKp strains, achieving accuracy, sensitivity, and specificity of 93%, 91%, and 96%, respectively [55].
The quantitative siderophore production test follows the same initial steps as the qualitative test. However, after incubation of the reaction mixture in the 96-well plate for 30 minutes in the dark, the results are read at 630 nm with a spectrophotometer, using a standard curve to quantify the siderophore concentration for each sample [138]. Although initially promising, a recent study by Russo et al has shown that the quantitative siderophore production was similar for ESBL-/CP hvKp and cKp (135.8 µg/mL vs. 139.8 µg/mL, respectively), indicating that: a) the test cannot reliably distinguish between contemporary hvKp and cKp strains, and b) it may be unsuitable for routine clinical laboratory use [33].
Mucoviscosity assay. Based on previous findings, the evaluation of mucoviscosity could also be used to distinguish between hvKp and cKp strains, given the stronger association of the hmv phenotype with the hypervirulent pathotype [139]. To assess mucoviscosity, according to Russo et al, cultures are grown in either Luria-Bertani (LB) or iron-chelated M9 minimum medium supplemented with trace elements (5 μg/mL CaCl2, 1 μg/mL CoCl2, 20 μg/mL MgCl2, 10 μg/mL MnCl2) (c-M9-CA-te) for 24 hours at 37°C. The OD600 is standardized to 1.0 using either LB or c-M9-te minimal medium (pre-spin OD600). After centrifugation of the OD600-adjusted cultures, 700 μL of supernatant are used for OD600 measurement (post-spin OD600). Mucoviscosity is recorded as the ratio of post-spin to pre-spin OD600 [33].
In their study, Russo et al found that in contrast to the quantitative siderophore production test, the mucoviscosity test was able to discriminate between ESBL-/CP hvKp and cKp (ie, the hvKp exhibited significantly higher mucoviscosity compared to cKp when grown in LB medium (P=0.0113) and in c-M9-CA-te medium (P< 0.0001)) [33].
Serum killing assay. Numerous studies have investigated the bactericidal activity of normal human serum against various Gram-negative bacteria [140], demonstrating minimal impact on some Kp strains [141]. Notably, the resistance to human serum could be a crucial VF.
According to Gao et al, 25 μL of bacterial suspension prepared from the mid-log phase is mixed with 75 μL of human serum and then incubated at 37°C for 3 hours. The response to serum killing is assessed in terms of viable cell counts and is graded on a scale from 1 to 6, where grades 1 to 2 represent high sensitivity, grades 3 to 4 indicate intermediate sensitivity, and grades 5 to 6 indicate resistance. Their findings revealed that 46.8% (22/47) of hvKp strains (all carrying rmpA/rmpA2 genes) demonstrated resistance to serum killing, with K1/K2 serotype strains exhibiting higher resistance compared to other serotypes [53]. Similarly, Wang et al corroborated this observation, indicating that K1/K2 serotypes displayed a higher resistance with grades 5 to 6 [131]. These results suggest that serum killing tests could be particularly useful for identifying hvKp of the K1/K2 types (Table 2).
Tellurite resistance test. Tellurite resistance in Kp is strongly associated with hvKp strains. The tellurite resistance gene (terW) is linked to the virulence of hvKp and is often found on the virulence plasmid pLVPK carried by hvKp [142]. However, it is not exclusive to hvKp [46, 55]. The tellurite resistance test is considered simple and easy. When strains form black colonies on selective media containing tellurite, they are presumptively considered to be hvKp strains [9].
Sanikhani et al screened a total of 477 non-repetitive Kp strains isolated from 2 educational hospitals in Iran between 2019 and 2020 using the tellurite resistance test. They found that 34.2% (163/477) of the isolates were tellurite resistant, and among these, 102 (62.6%) were defined as hvKp isolates based on PCR positivity for at least one among iucA, iutA, or peg-344 targets [143]. Furthermore, Wu et al established a MacConkey agar medium containing 4 μg/mL potassium tellurite to detect tellurite resistance. Testing a collection of cKp and hvKp strains (of which 13.3% and 70.6% positive for terW, respectively), this method showed accuracy, sensitivity and specificity of 94.9%, 92.7% and 100%, respectively. These results indicate that this test may be a promising adjuvant strategy to distinguish cKp from hvKp (Table 2). However, further confirmatory studies should be performed because presence of terW may not correspond to phenotypic tellurite resistance [144].
Multiplex quantitative PCR (multiplex qPCR). Quantitative PCR (qPCR) consists of the PCR amplification of a single or multiple target genes coupled with the quantitative detection of the exponentially amplified DNA product(s) by various methods, such as fluorescence emission with SYBR Green or TaqMan probes. It has emerged as a powerful tool in microbial diagnostics due to its high sensitivity, specificity, and rapidity of results [145, 146].
Xu et al established and developed a rapid quadruple qPCR assay that includes, in a single reaction, a positive control specific for Kp (gltA), 3 hvKp-specific markers (iucA, rmpA, and rmpA2), and 1 carbapenemase target (blaKPC) [15]. To validate the assay, a total of 84 Kp-containing clinical samples were tested showing that 31 had hvKp and 53 had cKp. Based on the outcomes of conventional PCR, it was confirmed that in the 31 hvKp the iucA gene was present in all strains, while other genes were distributed as follows: rmpA (n=20), rmpA2 (n=18), iroB (n=17), and peg-344 (n=1). Various combinations of these genes were also observed, with 8 out of 31 hvKp and 1 out of 53 cKp carrying blaKPC. From the clinical samples, 67 strains were successfully isolated and tested using the string test, resulting in 20 strains testing positive (hvKp) and 47 testing negative (cKp). There were 26 discrepant samples between the multiplex qPCR and the string test: 16 strains were identified as hvKp only by qPCR, and 10 strains were identified as hvKp only by the string test. Notably, in the 10 strains identified as hvKp by the string test, no VFs were detected by conventional PCR. To evaluate the accuracy of the multiplex qPCR, an outbred murine infection model was used (107 CFU inoculum), showing ~80% mortality rates in strains identified as hvKp by qPCR, while no deaths were observed in strains identified as cKp. These findings indicate that only the multiplex qRT-PCR assay can accurately distinguish hvKp from cKp strains (Table 2).
Loopmediated isothermal amplification (LAMP). The LAMP technique allows for the amplification and fluorescent detection of the target DNA at a constant temperature. Remarkably, a typical genomic extraction is not strictly required, as this highly sensitive method takes advantage of the strand displacement activity of the Bst DNA polymerase, which is robust to amplification inhibitors [147, 148].
In 2020, Liao et al designed a LAMP detection method for hvKp utilizing peg-344 as the target molecular biomarker. This approach achieved a sensitivity 100 times greater than that of conventional PCR to detect hvKp isolated from human blood samples. Their findings underscore the potential of LAMP technology for rapid molecular diagnostics of hvKp, given its affordability and simplicity of use [149]. However, we should emphasize that peg-344 is not always carried by hvKp strains (eg, the pLVPK virulence plasmid carries it, but not its variant pVir-CR-HvKp4) [11]. Therefore, the LAMP approach should target additional virulence genes.
The Eazyplex® Superbug CRE assay (Amplex Diagnostics, GmbH) is a commercial rapid LAMP assay designed to identify carbapenemases in Enterobacteriaceae, including Klebsiella spp. [150]. In combination with this test, Rödel et al aimed to investigate the performance of the Eazyplex® hvKp assay (Research-Use-Only) that specifically detects virulence genes in hvKp. This assay contains lyophilized master mixes featuring primers targeting rmpA/A2, iucC, iroC, ybt, and clb [151]. Their findings revealed that 14 out of 87 isolates from invasive infections (16.1%) harbored at least one of the virulence genes, with an increased Kleborate virulence score of ≥2 (see below). Among these, nine scored 4 or 5 (10.3%). The hypermucoviscosity assessed by the string test was positive for 7 of the 14 isolates, while rmpA/A2 were detected in 9 isolates. Of note, the time from test to result for this assay was less than 15 minutes, indicating that this assay can represent a valuable option for identifying hvKp, especially if used in combination with the Eazyplex® Superbug CRE assay to detect potential CR hvKp (Table 2) [151].
Recombinaseaided amplification (RAA). RAA is an isothermal amplification technique based on recombinase-aided polymerase amplification, known for its high sensitivity and specificity [9].
Yan et al established a rapid and convenient diagnostic tool for identifying infections caused by hvKp. The novel RAA assay targeting peg-344 and rmpA was validated on 208 clinical samples, including 158 Kp-positive samples collected from healthy individuals (n=60) and inpatients with pneumonia (n=80), bloodstream infection or liver abscess (n=68), demonstrating 100% sensitivity and specificity compared to qPCR by detecting all hvKp-positive samples, but owning a five-fold lower limit of detection [48].
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). MALDI-TOF MS is a rapid technique to discriminate unique protein signatures based on mass spectrometry [9].
In 2015, Huang et al developed a MALDI-TOF MS method for the detection and differentiation of K1 and non-K1 Kp strains with an accuracy of 94.1% and 90%, respectively [152]. This method is currently limited by the fact that it cannot distinguish hvKp with other capsule polysaccharide types. Therefore, it needs further improvement, but it could still be applied alongside other conventional genotyping techniques for its rapidity and accuracy (Table 2).
Immunochromatographic strip. Antigen detection by using immunochromatographic tests (ICTs) can be used to detect components of bacteria [153]. The ICTs are often lateral flow immune assays where antigen(s) recognition is assured by a monoclonal antibody (mAb) specific to the target analyte labelled with a visual tag. Results are simply interpreted by visualization of a colored line in the test pad. These ICTs are very useful because of their short execution time, low cost, accuracy, lack of additional instrumentation, ease of implementation and minimal hands-on time [153, 154].
Since the Kp serotypes K1 and K2 are primarily associated with community-acquired infections and bacteremia, and also often presenting the hvKp pathotype, Siu et al developed a colloidal-based immunochromatographic strip (ICS). This ICS incorporates anti-Kp capsular polysaccharide polyclonal antibodies and is designed to detect Kp serotypes K1 and K2 [155]. The ICS offers a rapid and straightforward detection method, providing results within 5 minutes after loading the samples obtained by placing diluted cultures onto the ICS. The assay was validated by testing 100 clinical isolates that had been collected from previous studies, including 30, 20, and 50 isolates of Kp serotypes K1, K2, and non-K1/K2, respectively. Notably, there were no false-positive or false-negative results observed with the ICS, when compared to the results obtained by conventional PCR and serum agglutination assay, indicating a high degree of sensitivity and specificity [155].
Wang et al aimed to further evaluate the efficacy of this ICS assay for detecting Kp serotypes K1 and K2 in pus samples from liver abscess and in positive blood culture samples [156]. Among the 108 Kp samples, ICS testing of blood culture samples identified the presence of 14 serotype K1 and 16 serotype K2 Kp. However, single colony testing revealed that 2 non-K1/K2 results were erroneously identified as serotype K2. PCR typing of these 2 isolates with discrepant results showed that both were of the K5 serotype [156].
As for the above MALDI-TOF MS approach, the ICS is currently limited by the fact that it can only detect Kp serotypes K1 and K2, and not specifically the hvKp pathotype (Table 2). However, this assay could be a valuable addition to current diagnostic methods for its rapidity and convenience.
Raman spectroscopy. The Raman spectroscopy is a rapid, low-cost, and highly-sensitive analytical technique where a scattered light is used to measure the vibrational energy modes of a sample to identify its chemical constituents. Specifically, this method can be used to identify bacteria and their ARGs or virulence genes [157]. For instance, Lu et al developed a convolutional neural network (machine learning algorithm) to interpret Raman spectra from 71 Kp strains showing good accuracy in identifying carbapenemase/mobile colistin resistance genes along with rmpA/rmpA2 [158]. More recently, several studies have shown the potency of Raman spectroscopy coupled by machine learning to identify hmv Kp strains. In this way, Fernandez-Manteca et al were able to identify with high accuracy (94%) the K1, K2, K54, and K57 capsular serotypes in 20 hmv Kp strains, also distinguishing 6 non-mucoid isolates [159]. In another study, Zhang et al differentiated 10 hmv from 10 cKp strains with 99% accuracy [160].
Overall, the Raman spectroscopy combined with artificial intelligence represents a very promising technique to rapidly identify hvKp strains. However, this technology remains distant from being implemented in clinical laboratories, with its application currently confined to research settings (Table 2).
The detection of Kp together with its pathotype and ARGs is crucial for the timely implementation of appropriate clinical interventions and infection control and prevention measures, particularly to promptly identify MDR hvKp. Due to the limitations of current detection methods in this regard (see above), there is an urgent need to develop new and more efficient diagnostic tools.
Next-generation sequencing (NGS) technologies. NGS provides a comprehensive, high-throughput approach to microbial genomic analysis by sequencing millions of DNA fragments at the same time, allowing a full characterization of the pathogen, including the identification of VFs and ARGs. Various NGS techniques, generating short (eg, Illumina and Ion Torrent) or long reads (eg, PacBio and Oxford Nanopore technology (ONT)) have been used for this scope (Table 2) [161].
In particular, there are many studies that have used short-read only NGS (short-NGS) for epidemiologic investigations, especially for genomic confirmation and characterization of hvKp isolates. For example, short-NGS has been implemented extensively to gain insight into hvKp suspect isolates (eg, string-test positive) from clinical specimens (eg, liver abscess) derived from retrospective studies, thereby further elucidating potential genomic mechanisms linked to the virulence phenotype (ie, presence of VFs) [99, 162–166]. However, despite the successful use of short-NGS to characterize hvKp isolates, it is well known that short-NGS may not accurately elucidate important genomic elements (eg, prmpA, prmpA2, and peg-344) that are typically localized in MGEs (ie, virulence plasmids). To overcome this problem, some studies have opted for long-read only NGS (long-NGS) to generate complete genomes of hvKp isolates using the ONT or PacBio platforms [167, 168]. Lastly, long-NGS using ONT, may be used to directly determine characteristic epigenetic signatures of hvKp. Ghosh et al showed that hvKp strains possess a significantly higher levels of methylation in chromosomal DNA and extrachromosomal elements compared to cKp; this hypermethylation (ie, GATC and CCWGG motifs) was particularly rich in the virulome rather than in genes not directly associated with virulence [169].
As discussed above, long-NGS allows for complete genome characterization of hvKp, making it an ideal choice for epidemiologic studies. In this context, it is worth noting that the MinION offered by ONT is a small portable sequencing device that allows the generation of long-reads generally in the range of >10 kb, thus capable of resolving structural variations, long repeat regions, and genomic copy-number alterations, requiring only a very little upfront capital investment [170].
Although PacBio reads have historically been more accurate (>99.9%) than ONT reads, newer basecalling models (eg, super accuracy (SUP) basecalling) allow the current Nanopore R10.4 technology (V14 chemistry) to offer comparable performance (>99.1%) [171, 172]. In addition, the availability of newer assembly polishing tools (eg, Medaka) further increases the usability of ONT data for single nucleotide polymorphism (SNP) studies.
For instance, the study by Foster-Nyarko et al utilized sequencing data obtained from 54 unique Kp strains to evaluate the efficacy and accuracy of ONT assemblies in identifying ARGs, STs, and VFs compared with short-NGS and hybrid methods [173]. The authors found that the implementation of the R9.4.1 flow cell generated basecalled data, implementing the Fast, High Accuracy, and SUP models, along with polishing assemblies with Medaka, produced high-quality assemblies for determining MLST, K/O locus type, VFs, and ARGs. However, the quality of such ONT assemblies was still not sufficient for SNP analyses (eg, outbreak investigations) compared to Illumina-only assemblies [173]. Therefore, higher-quality data generated by either newer Nanopore V14 chemistry or PacBio technology may be ideal for high-resolution Kp studies at the SNP level.
A more established approach is to combine both short-NGS and long-NGS to generate hybrid genome assemblies, appropriate for high-resolution SNP analyses, with the goal to accurately characterize and link ARGs and VFs to their corresponding genomic elements (ie, chromosome and plasmids) [174]. In this context, the implementation of hybrid assemblies has been important in large retrospective epidemiologic investigations of hvKp clinical isolates necessary to elucidate high-risk clones, ARGs, VFs, and associated genomic elements [4, 175–177]. Similarly, such high-resolution studies are also important for the characterization of MDR hvKp, especially those possessing carbapenemases, which may be plasmid-associated and thus of critical epidemiological importance [176, 178–180].
Regardless of the sequencing approach implemented, in silico screening for Kp VFs can be performed using specialized bioinformatics tools. For example, one of the best-known tools is Kleborate, which allows the user to detect VFs in a given genome assembly by assessing their presence in ICEKp (ybt, clb, iro, rmp) and virulence plasmids (iro, iuc, rmpA, rmpA2) [181]. In particular, Kleborate determines a virulence score ranging from 0 to 5 (ie, aerobactin, 3 points; colibactin, 1 point; yersiniabactin, 1 point), with a score of 3 or higher indicating significant VFs, which may help researchers differentiate hvKp from cKp at the genomic level, but not for predicting the hypervirulent pathotype [33, 163]. Lastly, Kleborate also provides further information such as Klebsiella spp. identification, ST and ARGs (including SNPs), K (capsule) and O antigen (LPS) serotype prediction (implementing Kaptive [182]), making it a powerful tool for Kp genomic investigations.
Other platforms, such as Pathogenwatch (https://pathogen.watch/) and Institute Pasteur (https://bigsdb.pasteur.fr/), both incorporating Kleborate, allow researchers to screen genome assemblies for Kp VFs. Finally, there are other (non-Kleborate-based) options for researchers to screen for VFs, such as AMRFinder, which implements the NCBI reference gene catalog database (https://www.ncbi.nlm.nih.gov/pathogens/refgene/), and the comprehensive virulence factor database (VFDB; http://www.mgc.ac.cn/VFs/) [183, 184], which mostly requires manual implementation, but can be used by other tools such as ABRicate (https://github.com/tseemann/abricate).
Metagenomics approaches. Metagenomic NGS (mNGS; or shotgun-metagenomic sequencing) is valuable for understanding the genetic diversity of a population of strains by analyzing capsular serotypes, identifying ARGs and virulence-associated genes directly from clinical specimens, unlike methods relying on traditional cultivation methods [161]. In this context, very few studies have implemented mNGS, specifically Illumina-based, to characterize hvKp directly from clinical specimens.
Liu et al detected a total of 30 metagenome reconstructed Kp strains implementing PanPhlAn (and companion StrainPhlAn) from 150 clinical specimens (the majority were sputa and bronchoalveolar lavage fluids). As a result, the authors identified 399 virulence-associated genes, showcasing the power of metagenomic sequencing to resolve Kp at the strain level and demonstrating the genetic diversity of virulence determinants found, including the ybt locus (yersiniabactin), iucABCD and iutA (aerobactin), iroBCDE and iroN (salmochelin), and the capsule production-associated genes (cpsA and rmpA) [185].
Case studies have also validated the use of mNGS to identify hvKp directly in clinical samples. For instance, in a study by Peng et al, one hvKp strain belonging to ST23-K1 with iutA and rmpA genes was successfully identified from bronchoalveolar lavage fluid [186]. Another case study by Xie et al rapidly identified one hvKp harboring rmpA/rmpA2 and iutA genes from a liver abscess drainage fluid sample [187]. Moreover, other exceptional cases have applied mNGS to identify hvKp concurrently or faster than culture-based methods [188, 189]. Finally, a case of the implementation of ONT-based mNGS was reported, which allowed the accurate and rapid identification (5-7 hours) of a hvKp in liver abscess fluid [190].
Therefore, the implementation of mNGS for the detection of hvKp from specimens obtained from a suspected site of infection is critical for clinical management, as it provides rapid and accurate pathogen detection, enabling personalized patient treatment strategies by overcoming the limitations of traditional culture-based methods (Table 2) [191].
The prevalence of hvKp strains has increased globally, posing a significant public health challenge worldwide. MDR hvKp strains (especially those producing ESBLs and/or carbapenemases) require rapid and accurate detection for effective clinical management. However, there are currently no agreed-upon biomarkers for this pathotype definition. Therefore, current conventional diagnostic methods, although simple to perform, still have many disadvantages, such as a limited specificity and sensitivity, resulting in an inability to identify hvKp strains with complete accuracy.
In vivo models are generally more accurate in assessing pathogen virulence. In this context, confirmation of the hypervirulent phenotype of Kp strains in the mouse model is crucial, especially when evaluating the accuracy of new tests developed to specifically detect hvKp. However, mice studies are time- and resource-consuming, while recent studies have highlighted the limitations of using the cheaper G. mellonella infection model as a reliable method to distinguish between hvKp and cKp strains. In contrast, other molecular in vitro methods, such as qPCR or LAMP-based tests, may exhibit superior sensitivity and specificity, especially when the exact biomarker targets used to define hvKp will be formally set.
Overall, there is an urgent need to develop new and rapid diagnostic methods in this field. NGS approaches, including technologies like ONT, able to simultaneously identify virulence genes and ARGs, have shown a great potential to become powerful tools for comprehensive pathogen characterization, but they still rely mostly on traditional culture isolation, affecting sensitivity and time to result (turnaround time). Conversely, methods like mNGS offer a promising avenue for molecular characterization directly from clinical specimens. Future advances in this area hold great promise for significantly improving diagnostic accuracy, guiding more effective treatment strategies, and improving infection control measures in clinical settings to prevent further large-scale spread of hvKp.
We thank the Swiss Government Excellence Scholarship for supporting Dania Al Ismail (ESKAS-NR: 2023.0325).
This work was supported by the Swiss National Science Foundation (SNF) grant No. 192514 (to AE) and by the SNF National Research Programme “Advancing 3R - Animals, Research and Society” (NRP79) grant No. 206400 (to AE).
None declared
1. Chang D, Sharma L, Dela Cruz CS, Zhang D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front Microbiol. 2021;12:750662. doi: 10.3389/fmicb.2021.750662. PubMed PMID: 34992583; PMCID: PMC8724557.
2. Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629-61. doi: 10.1128/MMBR.00078-15. PubMed PMID: 27307579; PMCID: PMC4981674.
3. Taraghian A, Nasr Esfahani B, Moghim S, Fazeli H. Characterization of Hypervirulent Extended-Spectrum b-Lactamase-Producing Klebsiella pneumoniae Among Urinary Tract Infections: The First Report from Iran. Infect Drug Resist. 2020;13:3103-11. doi: 10.2147/IDR.S264440. PubMed PMID: 32982325; PMCID: PMC7489934.
4. Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence. 2020;11(1):1215-24. doi: 10.1080/21505594.2020.1809322. PubMed PMID: 32921250; PMCID: PMC7549996.
5. Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8(7):1111-23. doi: 10.1080/21505594.2017.1317412. PubMed PMID: 28402698; PMCID: PMC5711391.
6. Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther. 2019;17(2):71-3. doi: 10.1080/14787210.2019.1555470. PubMed PMID: 30501374; PMCID: PMC6349525.
7. Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146(10):1913-6. PubMed PMID: 3532983.
8. Nataro JP. Pathogenesis--Thoughts from the Front Line. Microbiol Spectr. 2015;3(3). doi: 10.1128/microbiolspec.MBP-0012-2014. PubMed PMID: 26185071.
9. Zhang QB, Zhu P, Zhang S, Rong YJ, Huang ZA, Sun LW, Cai T. Hypervirulent Klebsiella pneumoniae detection methods: a minireview. Arch Microbiol. 2023;205(10):326. doi: 10.1007/s00203-023-03665-y. PubMed PMID: 37672079.
10. Torres L, Escorihuela A, Eslava A, Losada J, Avila A. Liver abscess caused by Klebsiella pneumoniae associated with septic endophthalmitis. Enferm Infecc Microbiol Clin. 1989;7(3):173-4. PubMed PMID: 2490689.
11. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3). doi: 10.1128/CMR.00001-19. PubMed PMID: 31092506; PMCID: PMC6589860.
12. Wang W, Tian D, Hu D, Chen W, Zhou Y, Jiang X. Different regulatory mechanisms of the capsule in hypervirulent Klebsiella pneumonia: “direct” wcaJ variation vs. “indirect” rmpA regulation. Front Cell Infect Microbiol. 2023;13:1108818. doi: 10.3389/fcimb.2023.1108818. PubMed PMID: 37180440; PMCID: PMC10168181.
13. Jin M, Jia T, Liu X, Yang M, Zhang N, Chen J, Yang X, Qin S, Liu F, Tang Y, Wang Y, Guo J, Chen Y, Li B, Wang C. Clinical and genomic analysis of hypermucoviscous Klebsiella pneumoniae isolates: Identification of new hypermucoviscosity associated genes. Front Cell Infect Microbiol. 2022;12:1063406. doi: 10.3389/fcimb.2022.1063406. PubMed PMID: 36683676; PMCID: PMC9846069.
14. Vandhana V, Saralaya KV, Bhat S, Shenoy Mulki S, Bhat AK. Characterization of Hypervirulent Klebsiella pneumoniae (Hv-Kp): Correlation of Virulence with Antimicrobial Susceptibility. Int J Microbiol. 2022;2022:4532707. doi: 10.1155/2022/4532707. PubMed PMID: 36032181; PMCID: PMC9410983.
15. Xu Z, Li B, Jiang Y, Huang J, Su L, Wu W, Pang Q, Li Z, Zhang J, Li X, Wang J, Cen F, Peng L, Liang J, Wang F, Liu C, Shen C, Liu Y, Yang Y. Development of a quadruple qRT-PCR assay for simultaneous identification of hypervirulent and carbapenem-resistant Klebsiella pneumoniae. Microbiol Spectr. 2024;12(1):e0071923. doi: 10.1128/spectrum.00719-23. PubMed PMID: 38059628; PMCID: PMC10783029.
16. Restuccia PA, Cunha BA. Klebsiella. Infect Control. 1984;5(7):343-7. doi: 10.1017/s0195941700060549. PubMed PMID: 6564087.
17. Magill SS, O’Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR, Emerging Infections Program Hospital Prevalence Survey T. Changes in Prevalence of Health Care-Associated Infections in U.S. Hospitals. N Engl J Med. 2018;379(18):1732-44. doi: 10.1056/NEJMoa1801550. PubMed PMID: 30380384; PMCID: PMC7978499.
18. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283-300. doi: 10.1111/joim.13007. PubMed PMID: 31677303; PMCID: PMC7057273.
19. Doshi S, Forbes JD, Mubareka S, Andany N. Disseminated hypervirulent Klebsiella pneumoniae causing endophthalmitis, and lung and liver abscesses. CMAJ. 2022;194(18):E645-E8. doi: 10.1503/cmaj.211413. PubMed PMID: 35534028; PMCID: PMC9259407.
20. Babouee Flury B, Dona V, Buetti N, Furrer H, Endimiani A. First two cases of severe multifocal infections caused by Klebsiella pneumoniae in Switzerland: characterization of an atypical non-K1/K2-serotype strain causing liver abscess and endocarditis. J Glob Antimicrob Resist. 2017;10:165-70. doi: 10.1016/j.jgar.2017.04.006. PubMed PMID: 28729207.
21. Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2016;16(9):e190-e5. doi: 10.1016/S1473-3099(16)30021-4. PubMed PMID: 27402393.
22. Sellick JA, Russo TA. Getting hypervirulent Klebsiella pneumoniae on the radar screen. Curr Opin Infect Dis. 2018;31(4):341-6. doi: 10.1097/QCO.0000000000000464. PubMed PMID: 29847328.
23. Zhang S, Zhang X, Wu Q, Zheng X, Dong G, Fang R, Zhang Y, Cao J, Zhou T. Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 2019;8:166. doi: 10.1186/s13756-019-0615-2. PubMed PMID: 31673355; PMCID: PMC6819602.
24. De Francesco MA, Tiecco G, Scaltriti E, Piccinelli G, Corbellini S, Gurrieri F, Crosato V, Moioli G, Marchese V, Foca E, Bertelli DA, Castelli F, Caruso A. First Italian report of a liver abscess and metastatic endogenous endophthalmitis caused by ST-23 hypervirulent Klebsiella pneumoniae in an immunocompetent individual. Infection. 2023;51(1):271-6. doi: 10.1007/s15010-022-01879-8. PubMed PMID: 35802342; PMCID: PMC9879799.
25. Lan P, Shi Q, Zhang P, Chen Y, Yan R, Hua X, Jiang Y, Zhou J, Yu Y. Core Genome Allelic Profiles of Clinical Klebsiella pneumoniae Strains Using a Random Forest Algorithm Based on Multilocus Sequence Typing Scheme for Hypervirulence Analysis. J Infect Dis. 2020;221(Suppl 2):S263-S71. doi: 10.1093/infdis/jiz562. PubMed PMID: 32176785.
26. Cubero M, Grau I, Tubau F, Pallares R, Dominguez MA, Linares J, Ardanuy C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin Microbiol Infect. 2016;22(2):154-60. doi: 10.1016/j.cmi.2015.09.025. PubMed PMID: 26454059.
27. Chen J, Zhang H, Liao X. Hypervirulent Klebsiella pneumoniae. Infect Drug Resist. 2023;16:5243-9. doi: 10.2147/IDR.S418523. PubMed PMID: 37589017; PMCID: PMC10426436.
28. Russo TA, Shon AS, Beanan JM, Olson R, MacDonald U, Pomakov AO, Visitacion MP. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One. 2011;6(10):e26734. doi: 10.1371/journal.pone.0026734. PubMed PMID: 22039542; PMCID: PMC3200348.
29. Liu Z, Gu Y, Li X, Liu Y, Ye Y, Guan S, Li J. Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med. 2019;39(2):167-75. doi: 10.3343/alm.2019.39.2.167. PubMed PMID: 30430779; PMCID: PMC6240523.
30. Li W, Sun G, Yu Y, Li N, Chen M, Jin R, Jiao Y, Wu H. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225-32. doi: 10.1093/cid/cit675. PubMed PMID: 24099919.
31. Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031-41. doi: 10.2147/IDR.S161075. PubMed PMID: 30104891; PMCID: PMC6074765.
32. Tang M, Kong X, Hao J, Liu J. Epidemiological Characteristics and Formation Mechanisms of Multidrug-Resistant Hypervirulent Klebsiella pneumoniae. Front Microbiol. 2020;11:581543. doi: 10.3389/fmicb.2020.581543. PubMed PMID: 33329444; PMCID: PMC7714786.
33. Russo TA, Alvarado CL, Davies CJ, Drayer ZJ, Carlino-MacDonald U, Hutson A, Luo TL, Martin MJ, Corey BW, Moser KA, Rasheed JK, Halpin AL, McGann PT, Lebreton F. Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. mBio. 2024;15(2):e0286723. doi: 10.1128/mbio.02867-23. PubMed PMID: 38231533; PMCID: PMC10865842.
34. Parrott AM, Shi J, Aaron J, Green DA, Whittier S, Wu F. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York City hospital through screening of virulence genes. Clin Microbiol Infect. 2021;27(4):583-9. doi: 10.1016/j.cmi.2020.05.012. PubMed PMID: 32461145.
35. Tian D, Wang M, Zhou Y, Hu D, Ou HY, Jiang X. Genetic diversity and evolution of the virulence plasmids encoding aerobactin and salmochelin in Klebsiella pneumoniae. Virulence. 2021;12(1):1323-33. doi: 10.1080/21505594.2021.1924019. PubMed PMID: 33970792; PMCID: PMC8115583.
36. Zhu J, Wang T, Chen L, Du H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front Microbiol. 2021;12:642484. doi: 10.3389/fmicb.2021.642484. PubMed PMID: 33897652; PMCID: PMC8060575.
37. Alvarez D, Merino S, Tomas JM, Benedi VJ, Alberti S. Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect Immun. 2000;68(2):953-5. doi: 10.1128/IAI.68.2.953-955.2000. PubMed PMID: 10639470; PMCID: PMC97229.
38. Llobet E, Campos MA, Gimenez P, Moranta D, Bengoechea JA. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect Immun. 2011;79(9):3718-32. doi: 10.1128/IAI.05226-11. PubMed PMID: 21708987; PMCID: PMC3165464.
39. Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073-8. doi: 10.1128/JCM.01924-13. PubMed PMID: 24088853; PMCID: PMC3838100.
40. Pan YJ, Lin TL, Chen CT, Chen YY, Hsieh PF, Hsu CR, Wu MC, Wang JT. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep. 2015;5:15573. doi: 10.1038/srep15573. PubMed PMID: 26493302; PMCID: PMC4616057.
41. Abbas R, Chakkour M, Zein El Dine H, Obaseki EF, Obeid ST, Jezzini A, Ghssein G, Ezzeddine Z. General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity. Biology (Basel). 2024;13(2). doi: 10.3390/biology13020078. PubMed PMID: 38392297; PMCID: PMC10886558.
42. Pan YJ, Fang HC, Yang HC, Lin TL, Hsieh PF, Tsai FC, Keynan Y, Wang JT. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol. 2008;46(7):2231-40. doi: 10.1128/JCM.01716-07. PubMed PMID: 18508935; PMCID: PMC2446917.
43. Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. PubMed PMID: 29209595; PMCID: PMC5702448.
44. Huang Y, Li J, Wang Q, Tang K, Cai X, Li C. Detection of carbapenem-resistant hypervirulent Klebsiella pneumoniae ST11-K64 co-producing NDM-1 and KPC-2 in a tertiary hospital in Wuhan. J Hosp Infect. 2023;131:70-80. doi: 10.1016/j.jhin.2022.09.014. PubMed PMID: 36183928.
45. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37-46. doi: 10.1016/S1473-3099(17)30489-9. PubMed PMID: 28864030.
46. Dai P, Hu D. The making of hypervirulent Klebsiella pneumoniae. J Clin Lab Anal. 2022;36(12):e24743. doi: 10.1002/jcla.24743. PubMed PMID: 36347819; PMCID: PMC9757020.
47. Chen Y, Chen Y. Clinical Challenges with Hypervirulent Klebsiella Pneumoniae (hvKP) in China. J Transl Int Med. 2021;9(2):71-5. doi: 10.2478/jtim-2021-0004. PubMed PMID: 34497746; PMCID: PMC8386330.
48. Yan C, Zhou Y, Du S, Du B, Zhao H, Feng Y, Xue G, Cui J, Gan L, Feng J, Fan Z, Fu T, Xu Z, Zhang Q, Zhang R, Cui X, Tian Z, Chen Y, Zhang T, Huang L, Yuan J. Recombinase-Aided Amplification Assay for Rapid Detection of Hypervirulent Klebsiella pneumoniae (hvKp) and Characterization of the hvKp Pathotype. Microbiol Spectr. 2023;11(2):e0398422. doi: 10.1128/spectrum.03984-22. PubMed PMID: 36912637; PMCID: PMC10100362.
49. Juan CH, Fang SY, Chou CH, Tsai TY, Lin YT. Clinical characteristics of patients with pneumonia caused by Klebsiella pneumoniae in Taiwan and prevalence of antimicrobial-resistant and hypervirulent strains: a retrospective study. Antimicrob Resist Infect Control. 2020;9(1):4. doi: 10.1186/s13756-019-0660-x. PubMed PMID: 31911832; PMCID: PMC6942382.
50. Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, Sepulveda VE, Quinn JD, Miller VL. A Klebsiella pneumoniae Regulatory Mutant Has Reduced Capsule Expression but Retains Hypermucoviscosity. mBio. 2019;10(2). doi: 10.1128/mBio.00089-19. PubMed PMID: 30914502; PMCID: PMC6437046.
51. Walker KA, Treat LP, Sepulveda VE, Miller VL. The Small Protein RmpD Drives Hypermucoviscosity in Klebsiella pneumoniae. mBio. 2020;11(5). doi: 10.1128/mBio.01750-20. PubMed PMID: 32963003; PMCID: PMC7512549.
52. Nassif X, Honore N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol. 1989;3(10):1349-59. doi: 10.1111/j.1365-2958.1989.tb00116.x. PubMed PMID: 2693894.
53. Gao Q, Shen Z, Qin J, Liu Y, Li M. Antimicrobial Resistance and Pathogenicity Determination of Community-Acquired Hypervirulent Klebsiella pneumoniae. Microb Drug Resist. 2020;26(10):1195-200. doi: 10.1089/mdr.2019.0439. PubMed PMID: 32354302.
54. Palmer LD, Skaar EP. Transition Metals and Virulence in Bacteria. Annu Rev Genet. 2016;50:67-91. doi: 10.1146/annurev-genet-120215-035146. PubMed PMID: 27617971; PMCID: PMC5125913.
55. Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56(9). doi: 10.1128/JCM.00776-18. PubMed PMID: 29925642; PMCID: PMC6113484.
56. Harada S, Doi Y. Hypervirulent Klebsiella pneumoniae: a Call for Consensus Definition and International Collaboration. J Clin Microbiol. 2018;56(9). doi: 10.1128/JCM.00959-18. PubMed PMID: 29950337; PMCID: PMC6113475.
57. Russo TA, MacDonald U, Hassan S, Camanzo E, LeBreton F, Corey B, McGann P. An Assessment of Siderophore Production, Mucoviscosity, and Mouse Infection Models for Defining the Virulence Spectrum of Hypervirulent Klebsiella pneumoniae. mSphere. 2021;6(2). doi: 10.1128/mSphere.00045-21. PubMed PMID: 33762316; PMCID: PMC8546679.
58. Namikawa H, Oinuma KI, Yamada K, Kaneko Y, Kakeya H, Shuto T. Predictors of hypervirulent Klebsiella pneumoniae infections: a systematic review and meta-analysis. J Hosp Infect. 2023;134:153-60. doi: 10.1016/j.jhin.2023.02.005. PubMed PMID: 36813165.
59. Sheng Z, Li J, Chen T, Zhu Y, Yu X, He X, Zheng Y, Ma C, Zheng M, Wang P, Li Z, Xu Y, Xie Q, Su Z, Chen S. Clinical and Microbiological Characteristics of Klebsiella pneumoniae Bloodstream Infection in a Chinese Hospital: Hypervirulent and Multiclonal. Infect Drug Resist. 2022;15:3981-90. doi: 10.2147/IDR.S371477. PubMed PMID: 35924022; PMCID: PMC9343176.
60. Wu J, Chen J, Wang Y, Meng Q, Zhao J. Siderophore iucA of hypermucoviscous Klebsiella pneumoniae promotes liver damage in mice by inducing oxidative stress. Biochem Biophys Rep. 2022;32:101376. doi: 10.1016/j.bbrep.2022.101376. PubMed PMID: 36340868; PMCID: PMC9634269.
61. Bailey DC, Alexander E, Rice MR, Drake EJ, Mydy LS, Aldrich CC, Gulick AM. Structural and functional delineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae. J Biol Chem. 2018;293(20):7841-52. doi: 10.1074/jbc.RA118.002798. PubMed PMID: 29618511; PMCID: PMC5961048.
62. Russo TA, Gulick AM. Aerobactin Synthesis Proteins as Antivirulence Targets in Hypervirulent Klebsiella pneumoniae. ACS Infect Dis. 2019;5(7):1052-4. doi: 10.1021/acsinfecdis.9b00117. PubMed PMID: 31032610; PMCID: PMC6625901.
63. Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10(1):77. doi: 10.1186/s13073-018-0587-5. PubMed PMID: 30371343; PMCID: PMC6205773.
64. Lam MMC, Wyres KL, Duchene S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. doi: 10.1038/s41467-018-05114-7. PubMed PMID: 30006589; PMCID: PMC6045662.
65. Shon AS, Russo TA. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol. 2012;7(6):669-71. doi: 10.2217/fmb.12.43. PubMed PMID: 22702521.
66. Alharbi MT, Almuhayawi MS, Nagshabandi MK, Tarabulsi MK, Alruhaili MH, Gattan HS, Al Jaouni SK, Selim S, Alanazi A, Alruwaili Y, Zaied SM, Faried OA. Antimicrobial Resistance Pattern, Pathogenicity and Molecular Properties of Hypervirulent Klebsiella pneumonia (hvKp) among Hospital-Acquired Infections in the Intensive Care Unit (ICU). Microorganisms. 2023;11(3). doi: ARTN 661 10.3390/microorganisms11030661. PubMed PMID: WOS:000958441200001.
67. Neumann B, Sturhof C, Rath A, Kieninger B, Eger E, Muller JU, von Poblocki A, Gerlitz N, Wollschlager P, Schneider-Brachert W, Schaufler K, Klaper K, Steinmann J. Detection and characterization of putative hypervirulent Klebsiella pneumoniae isolates in microbiological diagnostics. Sci Rep. 2023;13(1):19025. doi: 10.1038/s41598-023-46221-w. PubMed PMID: 37923898; PMCID: PMC10624845.
68. Lv J, Zhu J, Wang T, Xie X, Wang T, Zhu Z, Chen L, Zhong F, Du H. The Role of the Two-Component QseBC Signaling System in Biofilm Formation and Virulence of Hypervirulent Klebsiella pneumoniae ATCC43816. Front Microbiol. 2022;13:817494. doi: 10.3389/fmicb.2022.817494. PubMed PMID: 35464966; PMCID: PMC9019566.
69. El-Mahdy R, El-Kannishy G, Salama H. Hypervirulent Klebsiella pneumoniae as a hospital-acquired pathogen in the intensive care unit in Mansoura, Egypt. Germs. 2018;8(3):140-6. doi: 10.18683/germs.2018.1141. PubMed PMID: 30250833; PMCID: PMC6141227.
70. Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. Metabolite Transporter PEG344 Is Required for Full Virulence of Hypervirulent Klebsiella pneumoniae Strain hvKP1 after Pulmonary but Not Subcutaneous Challenge. Infect Immun. 2017;85(10). doi: 10.1128/IAI.00093-17. PubMed PMID: 28717029; PMCID: PMC5607406.
71. Dey T, Chakrabortty A, Kapoor A, Warrier A, Nag VL, Sivashanmugam K, Shankar M. Unusual Hypermucoviscous Clinical Isolate of Klebsiella pneumoniae with No Known Determinants of Hypermucoviscosity. Microbiol Spectr. 2022;10(3):e0039322. doi: 10.1128/spectrum.00393-22. PubMed PMID: 35647656; PMCID: PMC9241604.
72. Altayb HN, Elbadawi HS, Baothman O, Kazmi I, Alzahrani FA, Nadeem MS, Hosawi S, Chaieb K. Genomic Analysis of Multidrug-Resistant Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae Strain Lacking the Hypermucoviscous Regulators (rmpA/rmpA2). Antibiotics (Basel). 2022;11(5). doi: 10.3390/antibiotics11050596. PubMed PMID: 35625240; PMCID: PMC9137517.
73. Ali MR, Yang Y, Dai Y, Lu H, He Z, Li Y, Sun B. Prevalence of multidrug-resistant hypervirulent Klebsiella pneumoniae without defined hypervirulent biomarkers in Anhui, China: a new dimension of hypervirulence. Front Microbiol. 2023;14:1247091. doi: 10.3389/fmicb.2023.1247091. PubMed PMID: 37869673; PMCID: PMC10585048.
74. Dong N, Yang X, Chan EW, Zhang R, Chen S. Klebsiella species: Taxonomy, hypervirulence and multidrug resistance. EBioMedicine. 2022;79:103998. doi: 10.1016/j.ebiom.2022.103998. PubMed PMID: 35405387; PMCID: PMC9010751.
75. Zhao Q, Guo L, Wang LF, Zhao Q, Shen DX. Prevalence and characteristics of surgical site hypervirulent Klebsiella pneumoniae isolates. J Clin Lab Anal. 2020;34(9):e23364. doi: 10.1002/jcla.23364. PubMed PMID: 32424981; PMCID: PMC7521332.
76. Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26-34. doi: 10.1016/j.jgar.2021.02.020. PubMed PMID: 33667703.
77. Du FL, Huang QS, Wei DD, Mei YF, Long D, Liao WJ, Wan LG, Liu Y, Zhang W. Prevalence of Carbapenem-Resistant Klebsiella pneumoniae Co-Harboring blaKPC-Carrying Plasmid and pLVPK-Like Virulence Plasmid in Bloodstream Infections. Front Cell Infect Microbiol. 2020;10:556654. doi: 10.3389/fcimb.2020.556654. PubMed PMID: 33777826; PMCID: PMC7996060.
78. Yang X, Dong N, Liu X, Yang C, Ye L, Chan EW, Zhang R, Chen S. Co-conjugation of Virulence Plasmid and KPC Plasmid in a Clinical Klebsiella pneumoniae Strain. Front Microbiol. 2021;12:739461. doi: 10.3389/fmicb.2021.739461. PubMed PMID: 34819921; PMCID: PMC8606748.
79. Li G, Jia L, Wan L, Xia L, Gao A, Yang R, Sun R, Wang M, Du J, Lian X, Zhang R, Fang L, Liao X, Liu Y, Liu BT, Sun J. Acquisition of a novel conjugative multidrug-resistant hypervirulent plasmid leads to hypervirulence in clinical carbapenem-resistant Klebsiella pneumoniae strains. mLife. 2023;2(3):317-27. doi: 10.1002/mlf2.12086. PubMed PMID: 38817808; PMCID: PMC10989919.
80. Sattler J, Ernst CM, Zweigner J, Hamprecht A. High frequency of acquired virulence factors in carbapenemase-producing Klebsiella pneumoniae isolates from a large German university hospital, 2013-2021. Antimicrob Agents Chemother. 2024;68(11):e0060224. doi: 10.1128/aac.00602-24. PubMed PMID: 39365038.
81. Jia X, Zhu Y, Jia P, Liu X, Yu W, Li X, Xu Y, Yang Q. Emergence of a Superplasmid Coharboring Hypervirulence and Multidrug Resistance Genes in Klebsiella pneumoniae Poses New Challenges to Public Health. Microbiol Spectr. 2022;10(6):e0263422. doi: 10.1128/spectrum.02634-22. PubMed PMID: 36264236; PMCID: PMC9769819.
82. Huo B, Wei D, Huang Q, Huang S, Fan L, Li P, Qiu J, Ren Q, Wei C, Liu Y. Acquisition of a stable and transferable plasmid coharbouring hypervirulence and MDR genes with low fitness cost: Accelerating the dissemination of ST11-KL64 CR-HvKP. J Glob Antimicrob Resist. 2024;36:350-7. doi: 10.1016/j.jgar.2024.01.010. PubMed PMID: 38307249.
83. Jin L, Wang R, Gao H, Wang Q, Wang H. Identification of a Novel Hybrid Plasmid Encoding KPC-2 and Virulence Factors in Klebsiella pneumoniae Sequence Type 11. Antimicrob Agents Chemother. 2021;65(6). doi: 10.1128/AAC.02435-20. PubMed PMID: 33722891; PMCID: PMC8315960.
84. Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Wang R, Wang H. High Prevalence of Hypervirulent Klebsiella pneumoniae Infection in China: Geographic Distribution, Clinical Characteristics, and Antimicrobial Resistance. Antimicrob Agents Chemother. 2016;60(10):6115-20. doi: 10.1128/AAC.01127-16. PubMed PMID: 27480857; PMCID: PMC5038323.
85. Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. 2019;18(1):4. doi: 10.1186/s12941-018-0302-9. PubMed PMID: 30665418; PMCID: PMC6341648.
86. Khaertynov KS, Anokhin VA, Davidyuk YN, Nicolaeva IV, Khalioullina SV, Semyenova DR, Alatyrev EY, Skvortsova NN, Abrahamyan LG. Case of Meningitis in a Neonate Caused by an Extended-Spectrum-B-Lactamase-Producing Strain of Hypervirulent Klebsiella pneumoniae. Front Microbiol. 2017;8:1576. doi: 10.3389/fmicb.2017.01576. PubMed PMID: 28861076; PMCID: PMC5559536.
87. Su SC, Siu LK, Ma L, Yeh KM, Fung CP, Lin JC, Chang FY. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum b-lactamase. Antimicrob Agents Chemother. 2008;52(2):804-5. doi: 10.1128/AAC.01269-07. PubMed PMID: 18056273; PMCID: PMC2224741.
88. Xu M, Li A, Kong H, Zhang W, Chen H, Fu Y, Fu Y. Endogenous endophthalmitis caused by a multidrug-resistant hypervirulent Klebsiella pneumoniae strain belonging to a novel single locus variant of ST23: first case report in China. BMC Infect Dis. 2018;18(1):669. doi: 10.1186/s12879-018-3543-5. PubMed PMID: 30558549; PMCID: PMC6296127.
89. Campos-Madueno EI, Moser AI, Jost G, Maffioli C, Bodmer T, Perreten V, Endimiani A. Carbapenemase-producing Klebsiella pneumoniae strains in Switzerland: human and non-human settings may share high-risk clones. J Glob Antimicrob Resist. 2022;28:206-15. doi: 10.1016/j.jgar.2022.01.016. PubMed PMID: 35085791.
90. Lei TY, Liao BB, Yang LR, Wang Y, Chen XB. Hypervirulent and carbapenem-resistant Klebsiella pneumoniae: A global public health threat. Microbiol Res. 2024;288:127839. doi: 10.1016/j.micres.2024.127839. PubMed PMID: 39141971.
91. Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Cao B, Wang H. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553-60. doi: 10.1016/j.jinf.2015.07.010. PubMed PMID: 26304687.
92. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107-12. doi: 10.1016/j.ijid.2015.06.023. PubMed PMID: 26141415.
93. Wei DD, Wan LG, Deng Q, Liu Y. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis. 2016;85(2):192-4. doi: 10.1016/j.diagmicrobio.2015.03.012. PubMed PMID: 27049969.
94. Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, Lv J, Qi X, Hu L, Chen L, Kreiswirth BN, Zhang R, Pan J, Wang L, Yu F. Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates with Carbapenem Resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi: 10.3389/fcimb.2017.00182. PubMed PMID: 28560183; PMCID: PMC5432538.
95. Cejas D, Fernandez Canigia L, Rincon Cruz G, Elena AX, Maldonado I, Gutkind GO, Radice MA. First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J Clin Microbiol. 2014;52(9):3483-5. doi: 10.1128/JCM.00726-14. PubMed PMID: 25031447; PMCID: PMC4313191.
96. Yuan Y, Li Y, Wang G, Li C, Chang YF, Chen W, Nian S, Mao Y, Zhang J, Zhong F, Zhang L. blaNDM-5 carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrob Resist Infect Control. 2019;8:140. doi: 10.1186/s13756-019-0596-1. PubMed PMID: 31452874; PMCID: PMC6701021.
97. Shen Z, Gao Q, Qin J, Liu Y, Li M. Emergence of an NDM-5-Producing Hypervirulent Klebsiella pneumoniae Sequence Type 35 Strain with Chromosomal Integration of an Integrative and Conjugative Element, ICEKp1. Antimicrob Agents Chemother. 2019;64(1). doi: 10.1128/AAC.01675-19. PubMed PMID: 31611359; PMCID: PMC7187603.
98. Shu L, Dong N, Lu J, Zheng Z, Hu J, Zeng W, Sun Q, Chan EW, Zhou H, Hu F, Chen S, Zhang R. Emergence of OXA-232 Carbapenemase-Producing Klebsiella pneumoniae That Carries a pLVPK-Like Virulence Plasmid among Elderly Patients in China. Antimicrob Agents Chemother. 2019;63(3). doi: 10.1128/AAC.02246-18. PubMed PMID: 30559135; PMCID: PMC6395905.
99. Yang X, Sun Q, Li J, Jiang Y, Li Y, Lin J, Chen K, Chan EW, Zhang R, Chen S. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect. 2022;11(1):841-9. doi: 10.1080/22221751.2022.2049458. PubMed PMID: 35236251; PMCID: PMC8942559.
100. Li L, Li S, Wei X, Lu Z, Qin X, Li M. Infection with Carbapenem-resistant Hypervirulent Klebsiella Pneumoniae: clinical, virulence and molecular epidemiological characteristics. Antimicrob Resist Infect Control. 2023;12(1):124. doi: 10.1186/s13756-023-01331-y. PubMed PMID: 37953357; PMCID: PMC10642049.
101. Capitani V, Arcari G, Ambrosi C, Scribano D, Ceparano M, Polani R, De Francesco A, Raponi G, Ceccarelli G, Villari P, Palamara AT, Marzuillo C, Carattoli A. In vivo evolution to hypermucoviscosity and ceftazidime/avibactam resistance in a liver abscess caused by Klebsiella pneumoniae sequence type 512. mSphere. 2024;9(9):e0042324. doi: 10.1128/msphere.00423-24. PubMed PMID: 39171923; PMCID: PMC11423586.
102. Shi Q, Shen S, Tang C, Ding L, Guo Y, Yang Y, Wu S, Han R, Yin D, Hu F. Molecular mechanisms responsible KPC-135-mediated resistance to ceftazidime-avibactam in ST11-K47 hypervirulent Klebsiella pneumoniae. Emerg Microbes Infect. 2024;13(1):2361007. doi: 10.1080/22221751.2024.2361007. PubMed PMID: 38801099; PMCID: PMC11172257.
103. Liu N, Lou N, Huang J, Chen Z, Li B, Zhang Z, Hong Y, Cao L, Xiao W. Genomic surveillance indicates clonal replacement of hypervirulent Klebsiella pneumoniae ST881 and ST29 lineage strains in vivo. Front Microbiol. 2024;15:1375624. doi: 10.3389/fmicb.2024.1375624. PubMed PMID: 38440138; PMCID: PMC10910047.
104. Moore R, O’Shea D, Geoghegan T, Mallon PW, Sheehan G. Community-acquired Klebsiella pneumoniae liver abscess: an emerging infection in Ireland and Europe. Infection. 2013;41(3):681-6. doi: 10.1007/s15010-013-0408-0. PubMed PMID: 23381876.
105. Fazili T, Sharngoe C, Endy T, Kiska D, Javaid W, Polhemus M. Klebsiella pneumoniae Liver Abscess: An Emerging Disease. Am J Med Sci. 2016;351(3):297-304. doi: 10.1016/j.amjms.2015.12.018. PubMed PMID: 26992260.
106. Han X, Yao J, He J, Liu H, Jiang Y, Zhao D, Shi Q, Zhou J, Hu H, Lan P, Zhou H, Li X. Clinical and laboratory insights into the threat of hypervirulent Klebsiella pneumoniae. Int J Antimicrob Agents. 2024;64(3):107275. doi: 10.1016/j.ijantimicag.2024.107275. PubMed PMID: 39002700.
107. Li J, Ren J, Wang W, Wang G, Gu G, Wu X, Wang Y, Huang M, Li J. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(4):679-89. doi: 10.1007/s10096-017-3160-z. PubMed PMID: 29238932.
108. Lin YT, Liu CJ, Chen TJ, Fung CP. Long-term mortality of patients with septic ocular or central nervous system complications from pyogenic liver abscess: a population-based study. PLoS One. 2012;7(3):e33978. doi: 10.1371/journal.pone.0033978. PubMed PMID: 22479491; PMCID: PMC3313956.
109. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881-7. doi: 10.1016/S1473-3099(12)70205-0. PubMed PMID: 23099082.
110. Jung SW, Chae HJ, Park YJ, Yu JK, Kim SY, Lee HK, Lee JH, Kahng JM, Lee SO, Lee MK, Lim JH, Lee CH, Chang SJ, Ahn JY, Lee JW, Park YG. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol Infect. 2013;141(2):334-40. doi: 10.1017/S0950268812000933. PubMed PMID: 22578630; PMCID: PMC9152034.
111. Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decre D, Brisse S. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20(11):1812-20. doi: 10.3201/eid2011.140206. PubMed PMID: 25341126; PMCID: PMC4214299.
112. Liu C, Guo J, Fan S, Guo W, Qi H, Baker S, Du P, Cao B. An increased prevalence of carbapenem-resistant hypervirulent Klebsiella pneumoniae associated with the COVID-19 pandemic. Drug Resist Updat. 2024;77:101124. doi: 10.1016/j.drup.2024.101124. PubMed PMID: 39128195.
113. Yang Y, Liu JH, Hu XX, Zhang W, Nie TY, Yang XY, Wang XK, Li CR, You XF. Clinical and microbiological characteristics of hypervirulent Klebsiella pneumoniae (hvKp) in a hospital from North China. J Infect Dev Ctries. 2020;14(6):606-13. doi: 10.3855/jidc.12288. PubMed PMID: 32683351.
114. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107-18. doi: 10.4161/viru.22718. PubMed PMID: 23302790; PMCID: PMC3654609.
115. Chou A, Nuila RE, Franco LM, Stager CE, Atmar RL, Zechiedrich L. Prevalence of hypervirulent Klebsiella pneumoniae-associated genes rmpA and magA in two tertiary hospitals in Houston, TX, USA. J Med Microbiol. 2016;65(9):1047-8. doi: 10.1099/jmm.0.000309. PubMed PMID: 27392968; PMCID: PMC5068137.
116. Peirano G, Pitout JD, Laupland KB, Meatherall B, Gregson DB. Population-based surveillance for hypermucoviscosity Klebsiella pneumoniae causing community-acquired bacteremia in Calgary, Alberta. Can J Infect Dis Med Microbiol. 2013;24(3):e61-4. doi: 10.1155/2013/828741. PubMed PMID: 24421832; PMCID: PMC3852459.
117. European Centre for Disease Prevention and Control (ECDC). Emergence of hypervirulent Klebsiella pneumoniae ST23 carrying carbapenemase genes in EU/EEA countries, first update. 14 February 2024. ECDC: Stockholm; 2024.
118. Pomakova DK, Hsiao CB, Beanan JM, Olson R, MacDonald U, Keynan Y, Russo TA. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis. 2012;31(6):981-9. doi: 10.1007/s10096-011-1396-6. PubMed PMID: 21918907.
119. Fung CP, Lin YT, Lin JC, Chen TL, Yeh KM, Chang FY, Chuang HC, Wu HS, Tseng CP, Siu LK. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis. 2012;18(8):1322-5. doi: 10.3201/eid1808.111053. PubMed PMID: 22840473; PMCID: PMC3414011.
120. Bray AS, Zafar MA. Deciphering the gastrointestinal carriage of Klebsiella pneumoniae. Infect Immun. 2024;92(9):e0048223. doi: 10.1128/iai.00482-23. PubMed PMID: 38597634; PMCID: PMC11384780.
121. Yang J, Li Y, Tang N, Li J, Zhou J, Lu S, Zhang G, Song Y, Wang C, Zhong J, Xu J, Feng J. The human gut serves as a reservoir of hypervirulent Klebsiella pneumoniae. Gut Microbes. 2022;14(1):2114739. doi: 10.1080/19490976.2022.2114739. PubMed PMID: 36001493; PMCID: PMC9415575.
122. Chung DR, Lee H, Park MH, Jung SI, Chang HH, Kim YS, Son JS, Moon C, Kwon KT, Ryu SY, Shin SY, Ko KS, Kang CI, Peck KR, Song JH. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis. 2012;31(4):481-6. doi: 10.1007/s10096-011-1334-7. PubMed PMID: 21739348.
123. Raffelsberger N, Hetland MAK, Svendsen K, Smabrekke L, Lohr IH, Andreassen LLE, Brisse S, Holt KE, Sundsfjord A, Samuelsen O, Gravningen K. Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity. Gut Microbes. 2021;13(1):1939599. doi: 10.1080/19490976.2021.1939599. PubMed PMID: 34182896; PMCID: PMC8244762.
124. Mario E, Hamza D, Abdel-Moein K. Hypervirulent Klebsiella pneumoniae among diarrheic farm animals: A serious public health concern. Comp Immunol Microbiol Infect Dis. 2023;102:102077. doi: 10.1016/j.cimid.2023.102077. PubMed PMID: 37844369.
125. Zhang ZB, Lei L, Zhang HX, Dai HG, Song Y, Li L, Wang Y, Xia ZF. Molecular Investigation of Klebsiella pneumoniae from Clinical Companion Animals in Beijing, China, 2017-2019. Pathogens. 2021;10(3). doi: ARTN 271 10.3390/pathogens10030271. PubMed PMID: WOS:000633969900001.
126. Liu BT, Zhang XY, Wan SW, Hao JJ, Jiang RD, Song FJ. Characteristics of Carbapenem-Resistant Enterobacteriaceae in Ready-to-Eat Vegetables in China. Front Microbiol. 2018;9:1147. doi: 10.3389/fmicb.2018.01147. PubMed PMID: 29910786; PMCID: PMC5992273.
127. Li Y, Wang ZQ, Dong HY, Wang MZ, Qin SS, Chen S, Li RC. Emergence of tet(X4)-positive hypervirulent Klebsiella pneumoniae of food origin in China Lwt-Food Sci Technol. 2023;173. doi: ARTN 114280 10.1016/j.lwt.2022.114280. PubMed PMID: WOS:000908042000003.
128. Furlan JPR, Savazzi EA, Stehling EG. Genomic insights into multidrug-resistant and hypervirulent Klebsiella pneumoniae co-harboring metal resistance genes in aquatic environments. Ecotoxicol Environ Saf. 2020;201:110782. doi: 10.1016/j.ecoenv.2020.110782. PubMed PMID: 32497817.
129. Mohammed R, Nader SM, Hamza DA, Sabry MA. Occurrence of carbapenem-resistant hypervirulent Klebsiella pneumoniae in oysters in Egypt: a significant public health issue. Ann Clin Microbiol Antimicrob. 2024;23(1):53. doi: 10.1186/s12941-024-00711-5. PubMed PMID: 38886796; PMCID: PMC11184735.
130. Li G, Shi J, Zhao Y, Xie Y, Tang Y, Jiang X, Lu Y. Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur J Clin Microbiol Infect Dis. 2020;39(9):1673-9. doi: 10.1007/s10096-020-03890-z. PubMed PMID: 32318968.
131. Wang TC, Lin JC, Chang JC, Hiaso YW, Wang CH, Chiu SK, Fung CP, Chang FY, Siu LK. Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathog. 2021;13(1):40. doi: 10.1186/s13099-021-00439-z. PubMed PMID: 34154656; PMCID: PMC8218402.
132. Russo TA, MacDonald U. The Galleria mellonella Infection Model Does Not Accurately Differentiate between Hypervirulent and Classical Klebsiella pneumoniae. mSphere. 2020;5(1). doi: 10.1128/mSphere.00850-19. PubMed PMID: 31915230; PMCID: PMC6952204.
133. Mai D, Wu A, Li R, Cai D, Tong H, Wang N, Tan J. Identification of hypervirulent Klebsiella pneumoniae based on biomarkers and Galleria mellonella infection model. BMC Microbiol. 2023;23(1):369. doi: 10.1186/s12866-023-03124-0. PubMed PMID: 38030994; PMCID: PMC10685466.
134. Shi Q, Lan P, Huang D, Hua X, Jiang Y, Zhou J, Yu Y. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18(1):94. doi: 10.1186/s12866-018-1236-2. PubMed PMID: 30157774; PMCID: PMC6116568.
135. Yang Q, Jia X, Zhou M, Zhang H, Yang W, Kudinha T, Xu Y. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerg Microbes Infect. 2020;9(1):320-31. doi: 10.1080/22221751.2020.1721334. PubMed PMID: 32037975; PMCID: PMC7034084.
136. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697-705. doi: 10.1084/jem.20030857. PubMed PMID: 14993253; PMCID: PMC2213305.
137. Lee CH, Liu JW, Su LH, Chien CC, Li CC, Yang KD. Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis. 2010;14(8):e688-92. doi: 10.1016/j.ijid.2010.01.007. PubMed PMID: 20547084.
138. Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014;82(6):2356-67. doi: 10.1128/IAI.01667-13. PubMed PMID: 24664504; PMCID: PMC4019165.
139. Xu Q, Yang X, Chan EWC, Chen S. The hypermucoviscosity of hypervirulent K. pneumoniae confers the ability to evade neutrophil-mediated phagocytosis. Virulence. 2021;12(1):2050-9. doi: 10.1080/21505594.2021.1960101. PubMed PMID: 34339346; PMCID: PMC8331041.
140. Al-Busaidi B, Al-Muzahmi M, Al-Shabibi Z, Rizvi M, Al-Rashdi A, Al-Jardani A, Farzand R, Al-Jabri Z. Hypervirulent Capsular Serotypes K1 and K2 Klebsiella pneumoniae Strains Demonstrate Resistance to Serum Bactericidal Activity and Galleria mellonella Lethality. Int J Mol Sci. 2024;25(3). doi: 10.3390/ijms25031944. PubMed PMID: 38339222; PMCID: PMC10855873.
141. Short FL, Di Sario G, Reichmann NT, Kleanthous C, Parkhill J, Taylor PW. Genomic Profiling Reveals Distinct Routes To Complement Resistance in Klebsiella pneumoniae. Infect Immun. 2020;88(8). doi: 10.1128/IAI.00043-20. PubMed PMID: 32513855; PMCID: PMC7375759.
142. Passet V, Brisse S. Association of tellurite resistance with hypervirulent clonal groups of Klebsiella pneumoniae. J Clin Microbiol. 2015;53(4):1380-2. doi: 10.1128/JCM.03053-14. PubMed PMID: 25631812; PMCID: PMC4365200.
143. Sanikhani R, Moeinirad M, Solgi H, Hadadi A, Shahcheraghi F, Badmasti F. The face of hypervirulent Klebsiella pneumoniae isolated from clinical samples of two Iranian teaching hospitals. Ann Clin Microbiol Antimicrob. 2021;20(1):58. doi: 10.1186/s12941-021-00467-2. PubMed PMID: 34465335; PMCID: PMC8406009.
144. Wu X, Zhan F, Zhang J, Chen S, Yang B. Identification of hypervirulent Klebsiella pneumoniae carrying terW gene by MacConkey-potassium tellurite medium in the general population. Front Public Health. 2022;10:946370. doi: 10.3389/fpubh.2022.946370. PubMed PMID: 36091562; PMCID: PMC9448990.
145. Navarro E, Serrano-Heras G, Castano MJ, Solera J. Real-time PCR detection chemistry. Clin Chim Acta. 2015;439:231-50. doi: 10.1016/j.cca.2014.10.017. PubMed PMID: 25451956.
146. Sahoo R, Jadhav S, Nema V. Journey of technological advancements in the detection of antimicrobial resistance. J Formos Med Assoc. 2024;123(4):430-41. doi: 10.1016/j.jfma.2023.08.008. PubMed PMID: 37598038.
147. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. PubMed PMID: 10871386; PMCID: PMC102748.
148. Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): Expansion of its practical application as a tool to achieve universal health coverage. J Infect Chemother. 2020;26(1):13-7. doi: 10.1016/j.jiac.2019.07.020. PubMed PMID: 31519395.
149. Liao W, Long D, Huang Q, Wei D, Liu X, Wan L, Feng Y, Zhang W, Liu Y. Rapid Detection to Differentiate Hypervirulent Klebsiella pneumoniae (hvKp) From Classical K. pneumoniae by Identifying peg-344 With Loop-Mediated Isothermal Amplication (LAMP). Front Microbiol. 2020;11:1189. doi: 10.3389/fmicb.2020.01189. PubMed PMID: 32655515; PMCID: PMC7325879.
150. Sekowska A, Bogiel T, Gospodarek-Komkowska E. Evaluation of eazyplex(R) SuperBug CRE Test for Beta-Lactamase Genes Detection in Klebsiella spp. and P. aeruginosa Strains. Curr Microbiol. 2020;77(1):99-103. doi: 10.1007/s00284-019-01806-5. PubMed PMID: 31728697; PMCID: PMC6946722.
151. Rödel J, Pfeifer Y, Fischer MA, Edel B, Stoll S, Pfister W, Löffler B. Screening of Klebsiella pneumoniae Isolates for Carbapenemase and Hypervirulence-Associated Genes by Combining the Eazyplex(®) Superbug CRE and hvKp Assays. Antibiotics (Basel). 2023;12(6). doi: 10.3390/antibiotics12060959. PubMed PMID: 37370278; PMCID: PMC10294845.
152. Huang Y, Li J, Gu D, Fang Y, Chan EW, Chen S, Zhang R. Rapid Detection of K1 Hypervirulent Klebsiella pneumoniae by MALDI-TOF MS. Front Microbiol. 2015;6:1435. doi: 10.3389/fmicb.2015.01435. PubMed PMID: 26733976; PMCID: PMC4685062.
153. Boutal H, Moguet C, Pommies L, Simon S, Naas T, Volland H. The Revolution of Lateral Flow Assay in the Field of AMR Detection. Diagnostics (Basel). 2022;12(7). doi: 10.3390/diagnostics12071744. PubMed PMID: 35885647; PMCID: PMC9317642.
154. Andryukov BG. Six decades of lateral flow immunoassay: from determining metabolic markers to diagnosing COVID-19. AIMS Microbiol. 2020;6(3):280-304. doi: 10.3934/microbiol.2020018. PubMed PMID: 33134745; PMCID: PMC7595842.
155. Siu LK, Tsai YK, Lin JC, Chen TL, Fung CP, Chang FY. Development of a Colloidal Gold-Based Immunochromatographic Strip for Rapid Detection of Klebsiella pneumoniae Serotypes K1 and K2. J Clin Microbiol. 2016;54(12):3018-21. doi: 10.1128/JCM.01608-16. PubMed PMID: 27707941; PMCID: PMC5121394.
156. Wang CH, Lu PL, Liu EY, Chen YY, Lin FM, Lin YT, Chang FY, Lin JC. Rapid identification of capsular serotype K1/K2 Klebsiella pneumoniae in pus samples from liver abscess patients and positive blood culture samples from bacteremia cases via an immunochromatographic strip assay. Gut Pathog. 2019;11:11. doi: 10.1186/s13099-019-0285-x. PubMed PMID: 30828389; PMCID: PMC6385414.
157. Hassan M, Zhao Y, Zughaier SM. Recent Advances in Bacterial Detection Using Surface-Enhanced Raman Scattering. Biosensors (Basel). 2024;14(8). doi: 10.3390/bios14080375. PubMed PMID: 39194603; PMCID: PMC11352333.
158. Lu J, Chen J, Liu C, Zeng Y, Sun Q, Li J, Shen Z, Chen S, Zhang R. Identification of antibiotic resistance and virulence-encoding factors in Klebsiella pneumoniae by Raman spectroscopy and deep learning. Microb Biotechnol. 2022;15(4):1270-80. doi: 10.1111/1751-7915.13960. PubMed PMID: 34843635; PMCID: PMC8966003.
159. Fernandez-Manteca MG, Ocampo-Sosa AA, Vecilla DF, Ruiz MS, Roiz MP, Madrazo F, Rodriguez-Grande J, Calvo-Montes J, Rodriguez-Cobo L, Lopez-Higuera JM, Farinas MC, Cobo A. Identification of hypermucoviscous Klebsiella pneumoniae K1, K2, K54 and K57 capsular serotypes by Raman spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc. 2024;319:124533. doi: 10.1016/j.saa.2024.124533. PubMed PMID: 38820814.
160. Zhang LY, Tang JW, Tian BS, Huang Y, Liu XY, Zhao Y, Cui XX, Zhang XY, Qin YR, Li GH, Wang L. Identification of hypermucoviscous Klebsiella pneumoniae strains via untargeted surface-enhanced Raman spectroscopy. Anal Methods. 2024;16(42):7105-13. doi: 10.1039/d4ay01137f. PubMed PMID: 39234672.
161. Hilt EE, Ferrieri P. Next Generation and Other Sequencing Technologies in Diagnostic Microbiology and Infectious Diseases. Genes (Basel). 2022;13(9). doi: 10.3390/genes13091566. PubMed PMID: 36140733; PMCID: PMC9498426.
162. Merla C, Kuka A, Mileto I, Petazzoni G, Gaiarsa S, De Vitis D, Ardizzone M, Corbella M, Baldanti F, Cambieri P. One-year surveillance for hypervirulent Klebsiella pneumoniae detected carbapenem-resistant superbugs. Microbiol Spectr. 2024;12(3):e0329223. doi: 10.1128/spectrum.03292-23. PubMed PMID: 38289935; PMCID: PMC10913487.
163. Wahl A, Fischer MA, Klaper K, Muller A, Borgmann S, Friesen J, Hunfeld KP, Ilmberger A, Kolbe-Busch S, Kresken M, Lippmann N, Lubbert C, Marschner M, Neumann B, Pfennigwerth N, Probst-Kepper M, Rodel J, Schulze MH, Zautner AE, Werner G, Pfeifer Y. Presence of hypervirulence-associated determinants in Klebsiella pneumoniae from hospitalised patients in Germany. Int J Med Microbiol. 2024;314:151601. doi: 10.1016/j.ijmm.2024.151601. PubMed PMID: 38359735.
164. Liu S, Ding Y, Xu Y, Li Z, Zeng Z, Liu J. An outbreak of extensively drug-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a teaching hospital in Southwest China. Front Cell Infect Microbiol. 2022;12:979219. doi: 10.3389/fcimb.2022.979219. PubMed PMID: 36176583; PMCID: PMC9513609.
165. Raj S, Sharma T, Pradhan D, Tyagi S, Gautam H, Singh H, Sood S, Dhawan B, Das BK, Kapil A, Chaudhry R, Mohapatra S. Comparative Analysis of Clinical and Genomic Characteristics of Hypervirulent Klebsiella pneumoniae from Hospital and Community Settings: Experience from a Tertiary Healthcare Center in India. Microbiol Spectr. 2022;10(5):e0037622. doi: 10.1128/spectrum.00376-22. PubMed PMID: 36043878; PMCID: PMC9602566.
166. Du L, Zhang J, Liu P, Li X, Su K, Yuan L, Zhang Z, Peng D, Li Y, Qiu J. Genome sequencing and comparative genome analysis of 6 hypervirulent Klebsiella pneumoniae strains isolated in China. Arch Microbiol. 2021;203(6):3125-33. doi: 10.1007/s00203-021-02263-0. PubMed PMID: 33811489; PMCID: PMC8019302.
167. Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, Yang TC. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039-46. doi: 10.1093/jac/dky164. PubMed PMID: 29800340.
168. Macleod CK, Khokhar FA, Warne B, Wick R, Butcher R, Cassimon B, Hayden P, Holt K, Torok ME. Rapid Whole Genome Sequencing of Serotype K1 Hypervirulent Klebsiella pneumoniae from an Undocumented Chinese Migrant. Case Rep Infect Dis. 2021;2021:6638780. doi: 10.1155/2021/6638780. PubMed PMID: 34007495; PMCID: PMC8100418.
169. Ghosh D, Pal A, Mohapatra S, Raj S, Vivekanandan P. Distinct epigenetic signatures of classical and hypervirulent Klebsiella pneumoniae. mSphere. 2024;9(1):e0046423. doi: 10.1128/msphere.00464-23. PubMed PMID: 38112443; PMCID: PMC10826340.
170. Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348-65. doi: 10.1038/s41587-021-01108-x. PubMed PMID: 34750572; PMCID: PMC8988251.
171. Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, Topfer A, Alonge M, Mahmoud M, Qian Y, Chin CS, Phillippy AM, Schatz MC, Myers G, DePristo MA, Ruan J, Marschall T, Sedlazeck FJ, Zook JM, Li H, Koren S, Carroll A, Rank DR, Hunkapiller MW. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37(10):1155-62. doi: 10.1038/s41587-019-0217-9. PubMed PMID: 31406327; PMCID: PMC6776680.
172. Ni Y, Liu X, Simeneh ZM, Yang M, Li R. Benchmarking of Nanopore R10.4 and R9.4.1 flow cells in single-cell whole-genome amplification and whole-genome shotgun sequencing. Comput Struct Biotechnol J. 2023;21:2352-64. doi: 10.1016/j.csbj.2023.03.038. PubMed PMID: 37025654; PMCID: PMC10070092.
173. Foster-Nyarko E, Cottingham H, Wick RR, Judd LM, Lam MMC, Wyres KL, Stanton TD, Tsang KK, David S, Aanensen DM, Brisse S, Holt KE. Nanopore-only assemblies for genomic surveillance of the global priority drug-resistant pathogen, Klebsiella pneumoniae. Microb Genom. 2023;9(2). doi: 10.1099/mgen.0.000936. PubMed PMID: 36752781; PMCID: PMC9997738.
174. Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, Adam MP. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30(7):693-700. doi: 10.1038/nbt.2280. PubMed PMID: 22750884; PMCID: PMC3707490.
175. Kochan TJ, Nozick SH, Medernach RL, Cheung BH, Gatesy SWM, Lebrun-Corbin M, Mitra SD, Khalatyan N, Krapp F, Qi C, Ozer EA, Hauser AR. Genomic surveillance for multidrug-resistant or hypervirulent Klebsiella pneumoniae among United States bloodstream isolates. BMC Infect Dis. 2022;22(1):603. doi: 10.1186/s12879-022-07558-1. PubMed PMID: 35799130; PMCID: PMC9263067.
176. Turton JF, Payne Z, Coward A, Hopkins KL, Turton JA, Doumith M, Woodford N. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and ‘non-hypervirulent’ types ST147, ST15 and ST383. J Med Microbiol. 2018;67(1):118-28. doi: 10.1099/jmm.0.000653. PubMed PMID: 29205138.
177. He Z, Xu W, Zhao H, Li W, Dai Y, Lu H, Zhao L, Zhang C, Li Y, Sun B. Epidemiological characteristics an outbreak of ST11 multidrug-resistant and hypervirulent Klebsiella pneumoniae in Anhui, China. Front Microbiol. 2022;13:996753. doi: 10.3389/fmicb.2022.996753. PubMed PMID: 36212848; PMCID: PMC9537591.
178. Lorenzin G, Gona F, Battaglia S, Spitaleri A, Saluzzo F, Trovato A, di Marco F, Cichero P, Biancardi A, Nizzero P, Castiglione B, Scarpellini P, Moro M, Cirillo DM. Detection of NDM-1/5 and OXA-48 co-producing extensively drug-resistant hypervirulent Klebsiella pneumoniae in Northern Italy. J Glob Antimicrob Resist. 2022;28:146-50. doi: 10.1016/j.jgar.2022.01.001. PubMed PMID: 35017071.
179. Liu BT, Su WQ. Whole genome sequencing of NDM-1-producing serotype K1 ST23 hypervirulent Klebsiella pneumoniae in China. J Med Microbiol. 2019;68(6):866-73. doi: 10.1099/jmm.0.000996. PubMed PMID: 31107201.
180. Dong N, Lin D, Zhang R, Chan EW, Chen S. Carriage of blaKPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(12):3317-21. doi: 10.1093/jac/dky358. PubMed PMID: 30239821.
181. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188. doi: 10.1038/s41467-021-24448-3. PubMed PMID: 34234121; PMCID: PMC8263825.
182. Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2(12):e000102. doi: 10.1099/mgen.0.000102. PubMed PMID: 28348840; PMCID: PMC5359410.
183. Liu B, Zheng D, Zhou S, Chen L, Yang J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022;50(D1):D912-D7. doi: 10.1093/nar/gkab1107. PubMed PMID: 34850947; PMCID: PMC8728188.
184. Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021;11(1):12728. doi: 10.1038/s41598-021-91456-0. PubMed PMID: 34135355; PMCID: PMC8208984.
185. Liu J, Xu Z, Li H, Chen F, Han K, Hu X, Fang Y, Chen D. Metagenomic Approaches Reveal Strain Profiling and Genotyping of Klebsiella pneumoniae from Hospitalized Patients in China. Microbiol Spectr. 2022;10(2):e0219021. doi: 10.1128/spectrum.02190-21. PubMed PMID: 35319275; PMCID: PMC9045201.
186. Peng W, Wu Y, Lu R, Zheng Y, Chen J, Pan P. Successful treatment of acute respiratory distress syndrome caused by hypervirulent Klebsiella pneumoniae with extracorporeal membrane oxygenation and continuous renal replacement therapy: A case report and literature review. Front Med (Lausanne). 2022;9:936927. doi: 10.3389/fmed.2022.936927. PubMed PMID: 36091705; PMCID: PMC9449315.
187. Xie J, Zhu Z. A case report of pyogenic liver abscess caused by hypervirulent Klebsiella pneumoniae diagnosed by metagenomic next-generation sequencing. J Int Med Res. 2021;49(7):3000605211032793. doi: 10.1177/03000605211032793. PubMed PMID: 34315270; PMCID: PMC8323428.
188. Wan S, Zhou A, Chen R, Fang S, Lu J, Lv N, Wang C, Gao J, Li J, Wu W. Metagenomics next-generation sequencing (mNGS) reveals emerging infection induced by Klebsiella pneumoniaeniae. Int J Antimicrob Agents. 2024;63(2):107056. doi: 10.1016/j.ijantimicag.2023.107056. PubMed PMID: 38081548.
189. Li Y, Yang Y, Zheng Y, Gao Y, Shu G, Gai W, Guo Y, Deng X. Hypervirulent Klebsiella pneumoniae Mediated Hepatic Infarction Septic Shock After Rectal Cancer Surgery: A Case Report. Infect Drug Resist. 2024;17:1911-8. doi: 10.2147/IDR.S452705. PubMed PMID: 38766680; PMCID: PMC11102091.
190. Zhang T, Huang X, Xu T, Li S, Cui M. Pyogenic liver abscess caused by extended-spectrum b-lactamase-producing hypervirulent Klebsiella pneumoniae diagnosed by third-generation sequencing: a case report and literature review. J Int Med Res. 2023;51(10):3000605231206296. doi: 10.1177/03000605231206296. PubMed PMID: 37903314; PMCID: PMC10617275.
191. Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol. 2019;14:319-38. doi: 10.1146/annurev-pathmechdis-012418-012751. PubMed PMID: 30355154; PMCID: PMC6345613.
Submitted November 11, 2024 | Accepted January 8, 2025 | Published January 24, 2025
Copyright © 2025 The Authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License.