Grace Freeman-Gallant1,#, Kathleen McCarthy2, #, Jennifer Yates1,2, Karen Kulas1, Michael J. Rudolph3, David J. Vance1,2, and Nicholas J. Mantis1,2
1Division of Infectious Diseases, Wadsworth Center, New York State Department of Health,
Albany, New York
2Department of Biomedical Sciences, University at Albany, Albany, New York
3New York Structural Biology Center, New York, New York
#These authors contributed equally to this study with last names listed alphabetically
Nicholas J. Mantis
nicholas.mantis@health.ny.gov
Freeman-Gallant G, McCarthy K, Yates J, Kulas K, Rudolph MJ, Vance DJ, Mantis NJ. A Refined Human Linear B Cell Epitope Map of Outer Surface Protein C (OspC) From the Lyme Disease Spirochete, Borrelia burgdorferi. Pathogens and Immunity. 2025;10(1):159–186. doi: 10.20411/pai.v10i1.756
10.20411/pai.v10i1.756
Background: A detailed understanding of the human antibody response to outer surface protein C (OspC) of Borrelia burgdorferi has important implications for Lyme disease diagnostics and vaccines.
Methods: In this report, 13 peptides encompassing 8 reported OspC linear B-cell epitopes from OspC types A, B, and K, including the largely conserved C-terminus (residues 193-210), were evaluated by multiplex immunoassay (MIA) for IgG reactivity with ~700 human serum samples confirmed positive in a 2-tiered Lyme disease diagnostic assay (Bb+) and ~160 post-treatment Lyme disease (PTLD) serum samples. The vmp-like sequence E (VlsE) C6-17 peptide was included as a positive control.
Results: Serum IgG from Bb+ samples were reactive with 10 of the 13 OspC-derived peptides tested, with the C-terminal peptide (residues 193-210) being the most reactive. Spearman’s rank correlation matrices and hierarchical clustering revealed a strong correlation between 193-210 and VlsE C6-17 peptide reactivity but little demonstrable association between 193-210 and the other OspC peptides or recombinant OspC. OspC peptide reactivities (excluding 193-210) were strongly correlated with each other and were disproportionately influenced by a subset of pan-reactive samples. In the PTLD sample set, none of the OspC-derived peptides were significantly reactive over baseline, even though VlsE C6-17 peptide reactivity remained.
Conclusions: The asynchronous and potentially short-lived serologic response to OspC-derived peptides reveals the complexity of B-cell responses to B. burgdorferi lipoproteins and confounds interpretation of antibody profiles for Lyme disease diagnostics.
Lyme disease; antibody; epitope; vaccine; Borrelia burgdorferi; Borreliella; human; peptides
Lyme disease, also known as Lyme borreliosis, is the most common vector-borne infection in the United States, with an estimated 450,000 cases per year [1]. The primary etiologic agent of Lyme disease is the spirochetal bacterium, Borrelia burgdorferi sensu stricto1. In North America, B. burgdorferi is transmitted to humans by black-legged ticks, Ixodes scapularis and Ixodes pacificus. The spirochete proliferates at the site of the tick bite, typically resulting in an expanding skin lesion commonly referred to as erythema migrans [2–4]. In the absence of antibiotic intervention, B. burgdorferi can disseminate to peripheral tissues, organs, joints, and the central nervous system, potentially resulting in complications including neuroborreliosis, carditis, and/or Lyme arthritis weeks, months or even years later [2, 5]. Moreover, a small fraction of patients with Lyme disease who receive a full regimen of antibiotics will report persistent health issues (eg, fatigue, cognitive issues, musculoskeletal pain), a difficult to define syndrome termed post-treatment Lyme disease (PTLD) [6–9]. Developing rapid and accurate diagnostic tests capable of detecting and discriminating between LD, PTLD syndrome and B. burgdorferi reinfection are much needed [2, 10, 11].
The humoral response to B. burgdorferi is robust, resulting in detectable spirochete-specific B cells and serum IgM and IgG days and weeks following an infectious tick bite [12–15]. In patients with Lyme disease, antibodies are primarily directed against B. burgdorferi’s outer surface lipoproteins [16]. Among these, outer surface protein C (OspC; also known as BB_B19, P23 and P25) stands out. OspC is a ~21 kDa helical homodimeric lipoprotein expressed by B. burgdorferi during tick transmission and in the early stages of infection [17]. OspC is implicated in facilitating spirochete egress from the tick during the course of a blood meal, enabling survival in the early stages of mammalian skin infection, and modulating transmigration across vascular walls [17–24]. The highly immunogenic nature of OspC has lent itself to applications in Lyme disease serology-based diagnostics, including widely used commercial tests [25, 26]. For example, the peptide derived from the largely conserved C-terminus of OspC (peptide “C10”) is a component of the FDA-approved diagnostic test Borrelia VlsE1/pepC10 from Zeus Scientific [27–31]. In addition to their diagnostic utility, linear B-cell epitopes from different OspC types are a component of a widely used canine Lyme disease vaccine and are being considered for human use (see review article on this topic [32]).
While multiple linear human B-cell epitopes on OspC have been reported over the past 3 decades, differences in serologic assays, detection methodologies, and sample sizes from varying Lyme disease cohorts make it difficult to draw conclusions about relative reactivities of one peptide over another [28, 31–41]. This is problematic because determining the relationships between OspC-derived peptides and total OspC antibodies has practical implications for interpreting serologic assays. Moreover, there are >26 allelic variants or types of OspC reported in the United States, with amino acid sequence identities that range from 60% to 90% [42, 43]. As different OspC types are associated with varying degrees of virulence and invasiveness, tracking epitope-specific responses to those particular types is of paramount importance [44, 45]. In New York state, for example, OspC types A, B, and K, which are associated with more invasive disease, represent ~70% of all isolates [35, 45, 46]. With that in mind, we sought to validate human antibody reactivities to reported linear epitopes associated with OspC types A, B, and K to enable broad comparability of epitope usage across B. burgdorferi for diagnostic and vaccine development purposes.
1 It has been proposed that the Lyme disease spirochetes be reclassified as the genus Borreliella, although this proposal has not yet been widely accepted within the community. See: Adeolu M, Gupta RS. A phylogenomic and molecular marker-based proposal for the division of the genus Borrelia into 2 genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Van Leeuwenhoek. 2014 Jun;105(6):1049-1072. doi: 10.1007/s10482-014-0164-x. Epub 2014 Apr 18. PMID: 24744012. and Margos G, Marosevic D, Cutler S, Derdakova M, Diuk-Wasser M, Emler S, Fish D, Gray J, Hunfeldt KP, Jaulhac B, Kahl O, Kovalev S, Kraiczy P, Lane RS, Lienhard R, Lindgren PE, Ogden N, Ornstein K, Rupprecht T, Schwartz I, Sing A, Straubinger RK, Strle F, Voordouw M, Rizzoli A, Stevenson B, Fingerle V. There is inadequate evidence to support the division of the genus Borrelia. Int J Syst Evol Microbiol. 2017 Apr;67(4):1081-1084. doi: 10.1099/ijsem.0.001717. Epub 2017 May 5. Erratum in: Int J Syst Evol Microbiol. 2017 Jun;67(6):2073. doi: 10.1099/ijsem.0.002100. Hunfeldt, K-P (corrected to Hunfeld, K-P). PMID: 27930271.
Chemicals and reagents were obtained from Thermo Fisher Scientific, unless noted otherwise. Buffers were prepared by the Wadsworth Center’s Cell and Tissue Culture core facility.
Recombinant B. burgdorferi OspCA (residues 38 to 201; PDB ID 1GGQ; UniProt ID Q07337) [47], OspCB (residues 38 to 202; B. burgdorferi strain ZS7; PDB ID 7UJ2) [48] and OspCK (residues 38 to 202; B. burgdorferi strain 297; PDB ID 7UJ6) [49] were expressed in E. coli strain BL21 (DE3) and purified by nickel-affinity and size-exclusion chromatography, as described [50]. Recombinant OspCA with the C10 sequence (residues 201-210; PVVAESPKKP; “OspCA+C10”) was expressed and purified as above. Linear epitope prediction was done using Discotope 2.0 [51]. OspC peptides (>80% purity) as described in Table 1 were synthesized by Genemed Synthesis with a C-terminal GGGSK-biotin extension. We also synthesized the vmp-like sequence E (VlsE)-derived C6 B31-17 peptide (MKKDDQIAAAIALRGMA) with the GGGSK-biotin linker [52].
Table 1. OspC and VlsE-derived Peptides Used in This Study
|
#a |
Residues |
OspC type sequence (A, B, K) b |
IEDB ID c |
References |
|
1 |
9-33 |
ILMTLFLFISCNNSGKDGNTSANSA |
181204 |
|
|
2 |
40-55 |
A/B-PNLTEISKKITDSNAV |
559957 |
[39] |
|
3 |
71-86 |
A-EIAAKAIGKKIHQNNG |
12383 |
[81] |
|
4 |
132-146 132-146 133-147 |
A-ETFTNKLKEKHTDLG B-EEFSTKLKDNHAQLG K-EDFTKKLEGEHAQLG |
63756 ,745097 14380, 18025 |
[36] |
|
5 |
155-169 |
A-AKEAILKTNGTKTKG |
558968 181187 |
|
|
6 |
177-186 178-187 178-187 |
A-FESVEVLSKA B-SGSLESLSKA K-FKAVENLAKA |
NA |
[82] |
|
7 |
183-190d |
A/B-LSKAAKEM |
560173 |
[39] |
|
8 |
193-210 |
A-NSVKELTSPVVAESPKKP |
745122 |
|
|
C6 |
VlsE C6-17 |
MKKDDQIAAAIALRGMA |
41843 |
[52] |
a, Numbers correspond to peptides highlighted in Figure 1, except C6; b, The capital letters A, B, or K preceding the peptides indicate their OspC type sequence. c, Immune Epitope Database (IEDB.org) identifier. d, Residues 184-191 in Type B.
Commercial Lyme disease seronegative (Lot 10500586) and seropositive (Lot 10510438) pooled samples were used strictly as intra-assay and bead-coupling confirmation controls throughout this study (ACCURUN products 810 and 130, respectively; SeraCare). The Lyme disease seronegative samples (referred to as “controls” throughout the manuscript) consist of a commercial panel of 81 serum samples collected in 2017-2018 (Access Biologicals). Five of the 81 serum samples were classified as extreme outliers for VlsE C6-17 reactivity by interquartile range (falling outside Q3 + (3*IQR)) calculated using Microsoft Excel and therefore removed from the control panel. In the end, a total of 76 control samples were used as comparators with the diagnostic sample set, and 75 samples were used as comparators for the PTLD sample set as a result of sample availability.
B. burgdorferi seropositive (Bb+) serum samples (n = 696) were obtained from the Wadsworth Center’s Diagnostic Immunology Laboratory. The archived samples were previously subjected to 2-tiered testing consisting of (Tier 1) a C6 peptide screen (Immunetics; C6 Lyme ELISA) or Enzyme Linked Fluorescent Assay (ELFA; BioMerieux, VIDAS Lyme IgG II and Lyme IgM II) followed by (Tier 2) IgM and IgG detection by Western blot (MarDX; Trinity Biotech). B. burgdorferi-specific IgM reactivity was defined as ≥2 positive bands, with IgG reactivity defined as ≥5 positive bands. Serum samples were aliquoted, de-identified, and classified as IgM-positive/IgG-negative, IgM-positive/IgG-positive, or IgM-negative/IgG-positive, based on the Western blot results. Post-treatment Lyme disease (PTLD) serum samples (n=158) were kindly provided by the Lyme Disease Biobank at Nuvance Health® (Danbury, CT). PTLD was defined as described by Aucott [8].

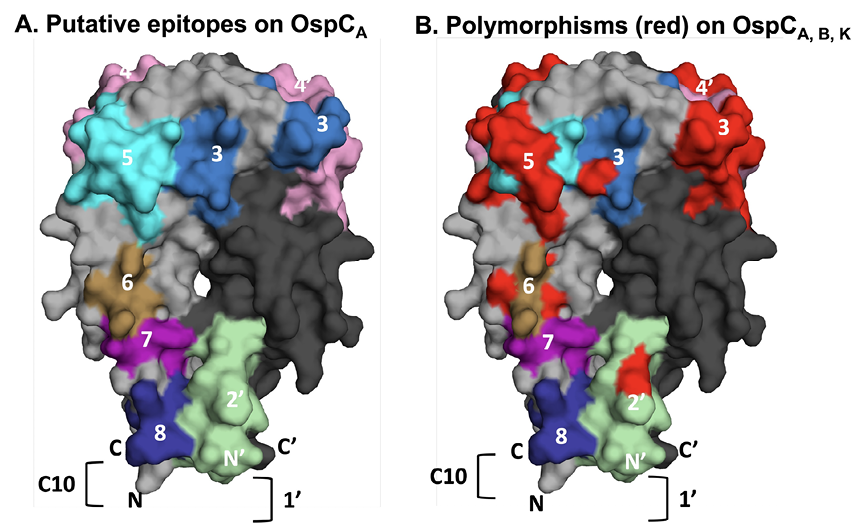
Figure 1. Relative location of linear epitopes on homodimeric OspCA. (A) Surface representation of homodimeric OspCA (residues 38-201; PDB 1GGQ] with one monomer (OspC) colored gray and the other, denoted with an apostrophe (OspC’), colored in charcoal. The linear epitopes examined in this study are coded and numbered according to Table 1. Epitope 1 is represented as a bracket (]) on OspC’, as its corresponding residues were truncated in the recombinant version of OspCA used for X-ray crystal structure analysis. Similarly, only residues 193-201 of epitope 8 (residues 193-210) are shown because the C-terminus of OspCA was truncated at position 201. Overlap between epitopes 6 and 7 are colored as epitope 6. (B) The same image as Panel A except that amino acid differences between OspC types A, B, and K within the 8 epitopes are colored red to illustrate degree of polymorphisms.
Recombinant OspCA, OspCB, or OspCK (5 μg) were coupled to Magplex-C microspheres (1 x 106) using sulfo-NHS (N-hydroxysulfosuccinimide) and EDC [1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride], as recommended by the manufacturer (Luminex Corp.). Coupled beads were diluted in xMap AbC Wash Buffer to a concentration of 5 x 106 beads/mL. Biotin-labeled peptides were complexed to Magplex-avidin microspheres following protocols provided by the manufacturer (Luminex Corp.). Microspheres were resuspended in 250 μL of assay buffer (1X PBS, 2% BSA, pH 7.4) then subjected to vortexing and sonication. A total of 1 x 106 beads in assay buffer was mixed with biotin-conjugated peptides (~5 μg) and incubated for 30 minutes at room temperature. The microsphere suspensions were then washed 3 times using wash buffer (1X PBS, 2% BSA, 0.02% Tween-20, 0.05% sodium azide, pH 7.4) and a magnetic separator, resuspended in 500 μL of assay buffer, and stored at 4oC until use. Successful coupling of OspC and peptides to the Magplex-C microspheres was confirmed by reactivity with immune serum and/or monoclonal antibodies.
For experimental use, assay buffer was used to dilute bead stocks (1:50) and human serum (1:100). The bead dilution (50 μL) and diluted serum (50 μL) were combined in black, clear-bottomed, non-binding, chimney 96-well plates (Greiner Bio-One) and allowed to incubate for 60 minutes in a tabletop shaker (600 rpm) at room temperature. Plates were washed 3 times using a magnetic separator and wash buffer. Secondary antibody goat anti-Human IgG Fc, eBioscience (Invitrogen) was diluted 1:500 in assay buffer and added (100 μL) to each well. The secondary antibody was allowed to incubate for 30 minutes in a tabletop shaker (600 rpm) at room temperature. Plates were washed as stated above and beads were resuspended in 100 μL of wash buffer. Samples were analyzed using a FlexMap 3D (Luminex Corp). To evaluate repeatability of the results, the entirety of the PTLD sample set was evaluated for peptide C6 and 193-210A reactivity at 2 different timepoints within the study. During these 2 evaluation time points beads were independently coupled, and results were generated months apart that demonstrated comparable results (data not shown). We defined sample positivity as the mean MFI + 6SD, based on previous studies conducted in the Wadsworth Center’s clinical laboratories [53, 54]. Index values were calculated using the sample MFI divided by the positivity cutoff for each bead set (ie, each peptide coated bead vs itself) such that an index values of >1.0 indicates positive reactivity for a given bead set.
For validation of the multiplex immunoassays (MIA) on an additional assay platform, enzyme-linked immunosorbent assays (ELISAs) were performed using a subset of the peptides and PTLD serum samples mentioned above. Nunc Maxisorb F96 microtiter plates (Thermo Fisher Scientific) were coated with rOspC or Streptavidin (0.1 μg/well) in PBS (pH 7.4), then incubated overnight at 4°C. The plates were washed 3 times with PBS-Tween 20 (PBS-T; 0.1%, vol/vol) and blocked with goat serum (2%, vol/vol, in PBS-T) for 2 hours at room temperature. For the plates to evaluate the peptides, biotinylated peptides were diluted (1.0 μg/well) and incubated for 1 hour. Plates were washed 3 times before being probed with serum samples (1:50 dilution). Plate-bound antibodies were detected with horseradish peroxidase (HRP)-labeled goat anti-human IgG polyclonal antibodies (SouthernBiotech). The plates were developed with 3,3, 5,5-tetramethylbenzidine (TMB; Kirkegaard & Perry Labs) and analyzed using a SpectraMax iD3 spectrophotometer and SoftMax version 7.1 (Molecular Devices).
Statistical analyses were performed using R 4.3.0 with R packages readxl and tidyverse [55–57]. MFIs (log10) were first subjected to the Shapiro-Wilk’s test to assess normality, then either Levene’s or Fligner-Killeen tests to compare variances. Mann-Whitney U tests (alpha level of 0.05) with Bonferroni P value adjustment were used to determine statistical significance for reactivity determination of OspC subtypes and OspC-derived peptides. R package corrplot [58] was used to generate the Spearman’s Rank correlation matrix. R package pheatmap was used to create the hierarchically clustered heatmaps [59]. R packages ggthemes (https://github.com/jrnold/ggthemes), RColorBrewer (https://cran.r-project.org/web/packages/RcolorBrewer/index.html), and ggpubr (https://CRAN.R-project.org/package=ggpubr) were used in formatting.
PyMol (PyMOL Molecular Graphics System, Version 3.0 Schrödinger, LLC) was used for epitope modeling using OspC PDB ID 1GGQ (strain B31, OspCA).
We sought to validate the reactivity of OspC linear B-cell epitopes, including those already accessioned in the Immune Epitope Database (iedb.org), that have been directly or indirectly implicated in Lyme disease diagnostics and/or B. burgdorferi immunity. Eight different epitopes, including a peptide encompassing the largely conserved C-terminus, were chosen for analysis (Table 1). When localized onto the structure of OspCA using PyMol, the epitopes represent ~30% of the surface area of the molecule (Figure 1A). An alignment of the primary amino acid sequences of the 3 OspC types (A, B, K) associated with invasive disease in the Northeast United States revealed polymorphisms in a number of these epitopes (Table 1; Figure 1B) [60, 61]. For this reason, a total of 13 different peptides were generated to encompass the specific amino acid sequences for each epitope within OspCA, OspCB, and OspCK (Table 1). The peptides were synthesized with a C-terminal biotin tag and coupled to streptavidin-coated microspheres for MIA by Luminex. As a positive control, we also coupled microspheres with the VlsE C6 B31-17 peptide from B. burgdorferi strain B31[52].
We had access to 2 B. burgdorferi seropositive (Bb+) serum sample collections for this study. A “diagnostic” collection consisting of 696 de-identified human serum samples from the Wadsworth Center’s Diagnostic Immunology (DI) laboratory previously classified as B. burgdorferi seropositive based on Western blot banding profiles (IgM+/IgG-, IgM+/IgG+, or IgM-/IgG+), as described in the Materials and Methods. A second collection provided by the Lyme Disease Biobank at Nuvance Health consisted of 158 serum samples from PTLD patients. These 2 collections were compared to a commercial panel consisting of ~75 serum samples confirmed negative for VlsE C6-17 peptide reactivity. We examined both IgG and IgM (see Supplemental Information) reactivity profiles.
The C-terminal residues of OspC are an immunodominant linear epitope used in several diagnostic assays [27–31]. However, there is some discrepancy in the literature as to what proportion C-terminus-specific antibodies constitute relative to total OspC antibodies. Some studies have reported that antibodies directed against the terminal 10 residues (C10) account for an overwhelming proportion of the total antibodies to OspC [62]. Others have suggested the opposite [63]. In an effort to resolve this question with our existing collection of Bb+ serum samples, we compared antibody reactivity to recombinant OspCA (rOspCA) with or without C-terminal residues 202-210 (VVAESPKKP). While MFIs for rOspCA with residues 202-210 were significantly greater than rOspCA without residues 202-210 in both the diagnostic and PTLD serum sample cohorts (Figure 2A-B), the results suggest that reactivity to the C-terminus constitutes only a fraction of the total antibody binding to rOspCA.
To address this issue further, we examined serum IgG antibody reactivity to OspC peptide 193-210A (which encompasses the conserved C-terminal amino acids) by Luminex and subjected the resulting MFI values to a linear regression model with MFIs obtained from rOspCA with or without C-terminus. Biotin-labeled OspC peptide 193-210A was coupled to Magplex-avidin microspheres then probed for IgG reactivity with both diagnostic and PTLD serum samples. Linear regression analysis revealed a weak association between OspC peptide 193-210A and rOspC reactivity, irrespective of the presence of the C-terminus (Figure 2B-F). This finding further supports the conclusion that C10-specific antibodies constitute only a small fraction of the total OspC reactivity.
The diagnostic and PTLD serum samples were next evaluated in MIA for reactivity with rOspC types A, B, K along with each of the 13 OspC-derived peptides listed in Table 1 in an effort to compare side-by-side relative binding to different peptide (linear) epitopes on OspC. The diagnostic serum IgG samples were significantly reactive with rOspC, with reactivity skewed towards OspCA and OspCB (Figure 3A). The diagnostic samples were also reactive with the 10 of the 13 OspC-derived peptides tested, with the 3 non-reactive peptides being 9-33ABK, 71-86A, and 178-187B (Figure 3A). The average serum IgG reactivity was highest against OspC peptide 193-210A and VlsE peptide C6-17 whose reactivities were significantly above controls (P<1x10-15 and P< 1x10-31, respectively). Analysis of IgM antibodies revealed significant reactivity with rOspC types A and B and peptides 9-33ABK and 178-187B, K but none of the other peptides tested (Supplementary Figure 1). Thus, our results confirm IgG reactivity with previously reported OspC linear epitopes.
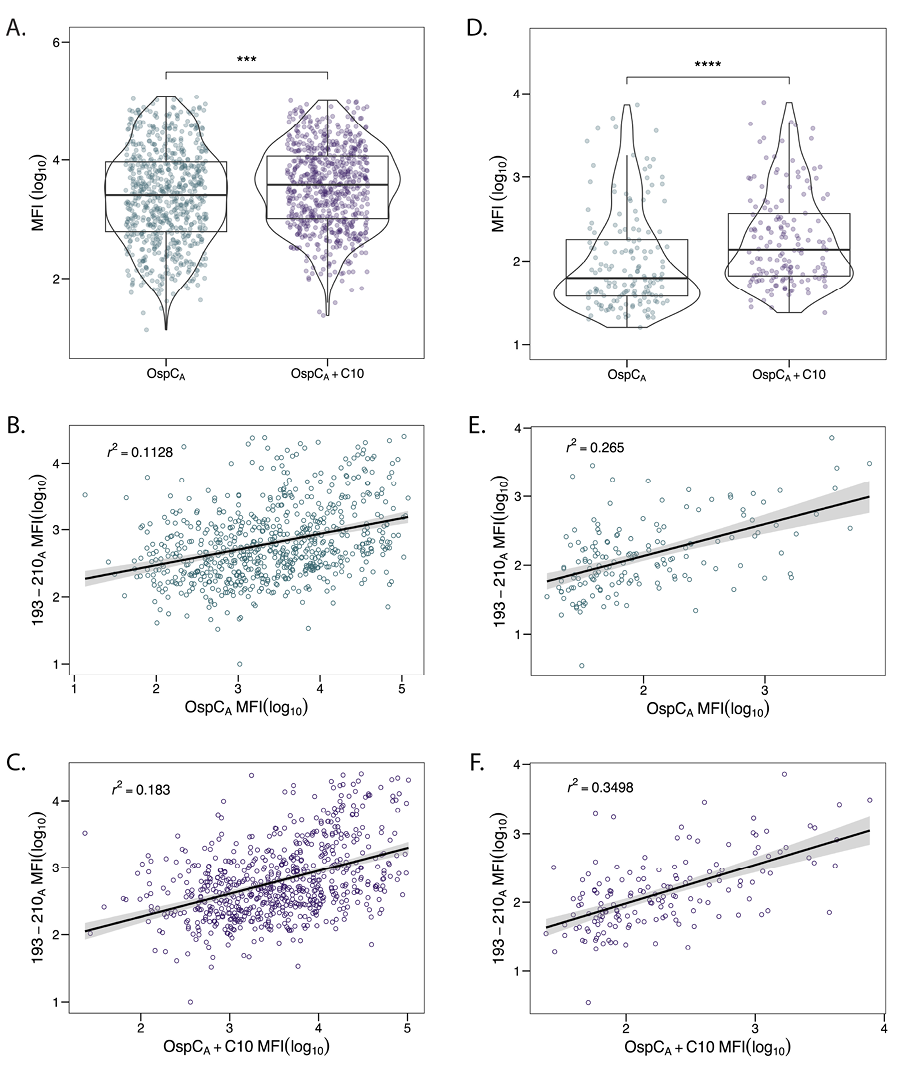
Figure 2. Correlation between OspCA reactivity and C10 region. Diagnostic (n=686) and PTLD (n=158) samples were subjected to Luminex with OspCA, OspCA+C10, and 193-210A as described in the Materials and Methods. OspCA MFIs were evaluated against OspCA+C10 for diagnostic (A) and PTLD (D) samples. Significance was determined by Mann-Whitney U test. *, P < 0.05. To examine the contribution of the C10 region to OspCA reactivity, MFIs for peptide 193-210A were plotted (on the y-axis) versus MFIs from OspCA and OspCA+C10 (on the x-axis) for diagnostic samples (B, C) and PTLD samples (E, F). Best fit lines and confidence intervals (level = 0.95) were added to each plot using a linear regression model. Adjusted R-squared (r2) values were calculated in R.
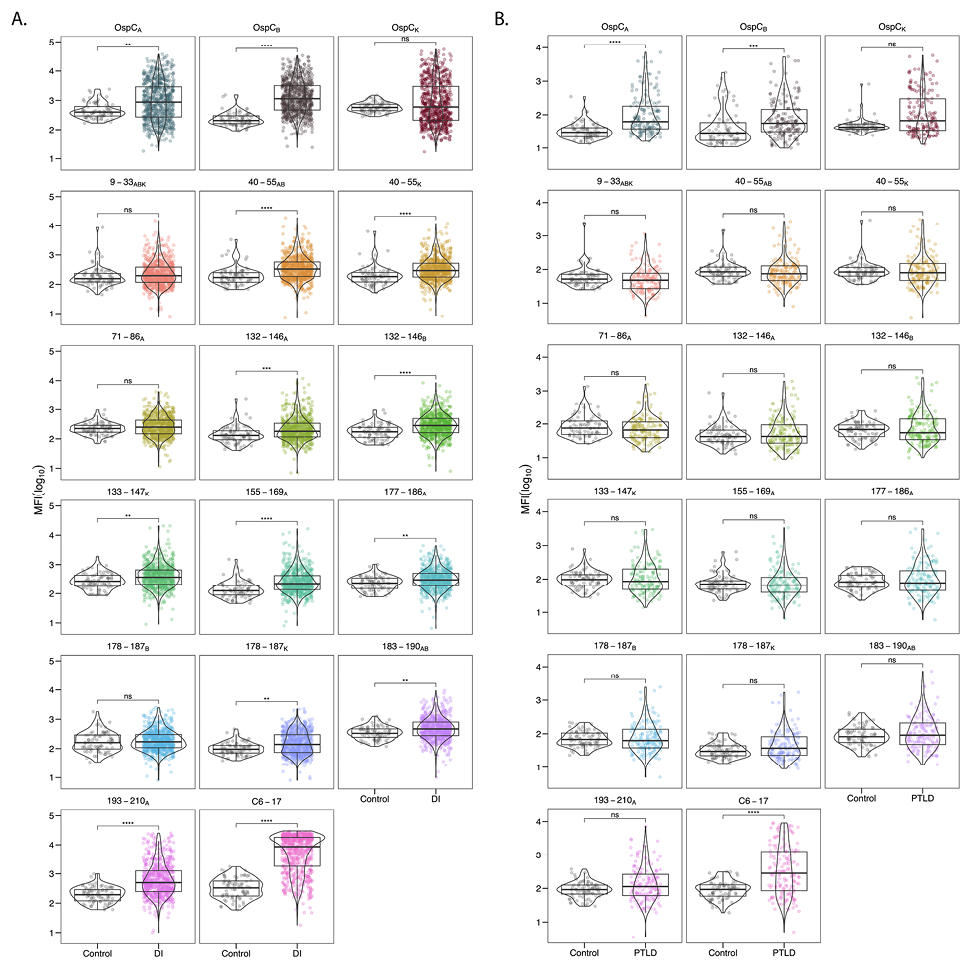
Figure 3. Serum IgG reactivity of diagnostic and PTLD samples with OspC and OspC-derived peptides. (A) Diagnostic and (B) PTLD serum samples (diluted 1:100) were subject to MIA using microspheres coated with recombinant dimeric OspC types A, B, and K (top rows), OspC-peptides as described in Table 1, and C6-17 peptide. Panels are labeled by corresponding residue numbers for each OspC type. MFI values were log10 transformed and compared to the control sample set. Significance was determined by the Mann-Whitney U-test with Bonferroni P-value adjustment (*, P< 0.05).
From the standpoint of using peptides as diagnostics for Lyme disease, we were particularly interested in peptides that had widespread reactivity within the known Bb+ cohorts and high MFI values relative to healthy controls. Garnering such information from previous studies is difficult due to relatively small sample sizes (see references cited in Table 1). To accomplish this, MFI values were subjected to a cutoff value of >6SD above the mean of the control MFI, log10 transformed, then plotted to reveal both the number of serum samples above that cutoff (% positive) as well as the magnitude (fold increase) of values over the cutoff. We also derived box and whisker plots to illustrate the upper and lower quartiles as well as the median reactivity to each antigen/peptide with superimposed violin plots to outline sample distribution. Using these metrics, reactivity with OspCA, OspCB, and OspCK ranged from 24% to 42% of the diagnostic samples with the fold increase ranging from 2.8 to 4.7 (Table 2; Figure 4A).
In terms of peptides, VlsE C6-17 was the most reactive (71% positive; fold increase 5.8), followed by 193-210A (26% positive; fold increase 4.4) (Table 2; Figure 4B). The percentage of samples >6SD for the other 12 peptides ranged from 0.1% to 10% with the fold increase between 1.2 and 2.5 (Table 2; Figure 4B). Lowering the cutoff threshold to >3SD resulted in higher overall peptide reactivities, as expected, but the pattern remained the same as >6SD (Supplementary Table 1). Thus, within a given cohort of B. burgdorferi seropositive samples, high serum IgG reactivity to OspC-derived peptides (apart from 193-210A) is limited to a small subset of individuals, which has important consequences for use of such peptides in diagnostic applications. In terms of IgM reactivity, the trends were similar to IgG except that the most pronounced antibody interactions were with peptides 177-186B, K followed by 9-33ABK (Supplementary Table 2). The differential peptide reactivity profiles between IgG and IgM may have diagnostic utility.
The same analysis was performed with the panel of PTLD samples. The PTLD samples were highly reactive with OspCA and OspCB with 16% and 18% of the samples having MFIs >6SD cutoff and fold increases between 2.8 and 5 (Table 2; Figures 3B, 4C). In the case of OspCK, only 3.2% were above the cutoff with a fold increase of 2.1 (Table 2; Figure 4C). In terms of linear epitopes, reactivity significantly above controls was limited to VlsE C6-17 (40% positive; fold increase 5.6) (Table 2: Figure 3B; Figure 4D). Only a small fraction of the PTLD serum samples were found to be positive for the 13 OspC peptides (range 0% to 14%) and the MFIs did not achieve statistical significance (Table 2; Figure 3B). PTLD samples were plotted to visualize OspC and peptide reactivities relative to the cutoff (Figure 4C-D). There is notably less reactivity to both 193-210A and C6-17 in the PTLD cohort in comparison to the diagnostic serum set.
Table 2. Significant IgG reactivity (>6SD) to OspCA/B/K and OspC peptides in diagnostic and PTLD serum samples
|
Diagnostic |
PTLD |
|||
|
OspC or peptide |
n = (%)a |
Fold increase (SD) b |
n = (%)a |
Fold increase (SD) b |
|
OspCA |
167 (24.0) |
2.8 (1.9) |
29 (18.4) |
5.5 (5.6) |
|
OspCB |
287 (41.2) |
3.7 (3.4) |
5 (3.2) |
2.1 (0.6) |
|
OspCK |
195 (28.0) |
4.7 (4.3) |
26 (16.5) |
2.8 (2.2) |
|
9-33ABK |
1 (0.1) |
2.4 (N/A) |
0 (0) |
N/A (N/A) |
|
40-55AB |
17 (2.4) |
2.1 (1.4) |
4 (2.5) |
1.6 (0.5) |
|
40-55K |
7 (1.0) |
1.5 (0.5) |
2 (1.3) |
1.3 (0.2) |
|
71-86A |
28 (4.0) |
1.7 (0.8) |
1 (0.6) |
1.4 (NA) |
|
132-146A |
18 (2.6) |
2.4 (1.5) |
5 (3.2) |
1.7 (0.6) |
|
132-146B |
48 (6.9) |
2.0 (1.3) |
11 (7.0) |
3.0 (1.9) |
|
133-147K |
36 (5.2) |
2.4 (2.6) |
8 (5.1) |
2.5 (1.2) |
|
155-169A |
30 (4.3) |
2.5 (2.5) |
5 (3.2) |
2.5 (1.5) |
|
177-186A |
38 (5.5) |
1.6 (0.7) |
13 (8.2) |
2.6 (2.0) |
|
178-187K |
70 (10.1) |
1.5 (0.7) |
15 (9.5) |
3.0 (2.8) |
|
178-187B |
7 (1.0) |
1.2 (0.1) |
12 (7.6) |
2.7 (1.9) |
|
183-190ABc |
53 (7.6) |
1.9 (1.0) |
14 (8.9) |
3.0 (3.1) |
|
193-210A (C10) |
185 (26.6) |
4.4 (4.5) |
22 (14.0) |
3.3 (3.0) |
|
VlsE C6-17 |
496 (71.3) |
5.8 (2.9) |
64 (40.5) |
5.6 (5.0) |
a, The number of samples (n =) and percent reactivity (%) to each antigen in the Diagnostic (n=696) and PTLD (n=158) sample sets were calculated using a cutoff of >6SD above the mean of control samples; b, The fold increase (magnitude) of positive sample antibody reactivity over controls such that an index value of 1 is 6SD above the control mean. For example, 2.8 corresponds to ~16SD greater than the mean control. c, OspCB residues 184-191.
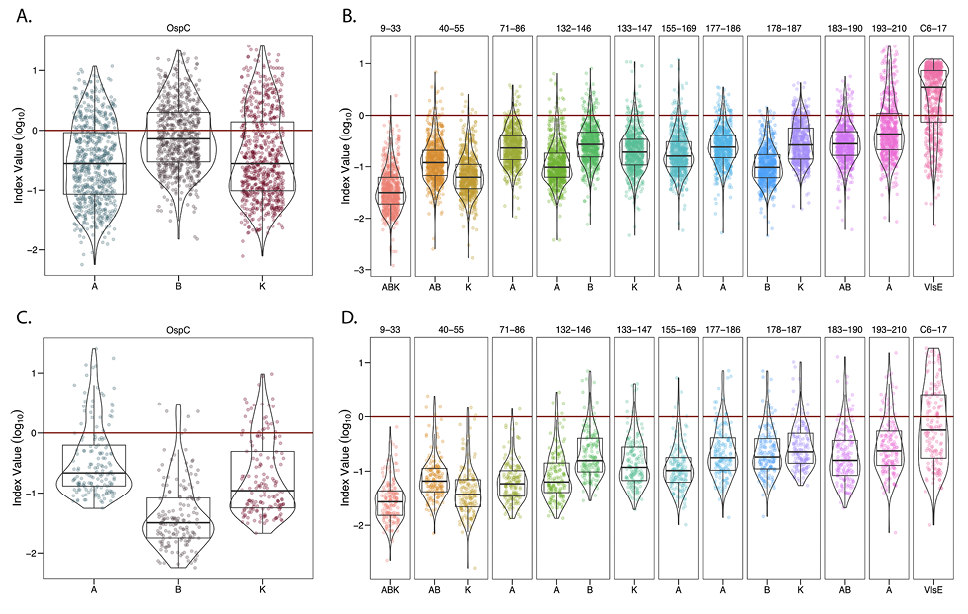
Figure 4. Indexed (>6SD) IgG reactivity of DI and PTLD serum samples with OspC and OspC-derived peptides. MFI values for DI (A, B) and PTLD (C, D) serum samples reactive with rOspC (left panels A, C) and OspC- and VlsE-derived peptides (right panels B, D) were indexed using a cutoff of 6SD above mean control reactivity. The resulting values were then log10 transformed with each point representing a single sample. OspC subtypes are listed along the x-axis, residues are indicated along the top of the plot, and each antigen is represented by a different color. In each of the graphs, the 6SD cutoff corresponds to a value of 0 on the y-axis and is depicted with horizontal red line. All samples falling above y = 0 were found to be positive for the given antigen and were used to calculate percentage positivity and fold increase as reported in Tables 2 and 3. Box and whisker plots illustrate the upper and lower quartiles as well as the median reactivity to each antigen (thick center line). Sample points that fall above or below the whiskers are considered outliers. Violin plots outline the distribution of data.
We postulated that antibody reactivity with the OspC-derived peptides would correlate with overall OspC antibody levels in any given sample, assuming that the 2 antibody populations track with each other and serve as redundant indicators of a previous B. burgdorferi infection. To investigate these relationships, we generated correlation matrices (Spearman’s Rank) assessing the correlation between OspC and each of the different OspC-derived peptides, including 193-210A, and VlsE C6-17 (Figure 5). While all correlations were determined to be positive, peptide 193-210A reactivity was only weakly correlated with OspCA, OspCB, or OspCK reactivity (rs = 0.2-0.4) in the diagnostic samples (Figure 5A). Rather, 193-210A reactivity was moderately to strongly correlated with other OspC-derived peptides, including 132-146A (rs = 0.63), 183-190AB (rs = 0.58), and 40-55ABK (rs = 0.58). Indeed, inter-peptide correlations were generally strong across the board (eg, 132-146B versus 177-186A, rs > 0.9). A similar pattern was observed in the PTLD samples: OspC-derived peptide reactivities were strongly correlated with each other (rs = 0.63-0.98), but weakly correlated with OspCA, OspCB, and OspCK (rs = 0.2-0.4) (Figure 5B). In the case of VlsE C6-17, reactivity was weakly correlated with OspC and OspC-derived peptides in both the diagnostic and PTLD samples, except for moderate correlativity with 193-210A (C10) in the PTLD cohort (rs = 0.56). These results reveal a significant degree of discordance (asynchrony) among circulating antibody levels against OspC, OspC-derived peptides, and the VlsE C6-17 peptide in both diagnostic and PTLD cohorts.
The absence of a strong correlation between antibody levels to OspC and OspC-derived peptides led us to subject the diagnostic and PTLD datasets to hierarchical clustering (HC) as a means to identify possible relationships in immunoreactivity profiles. In the case of the diagnostic samples, the horizontal (top) dendrogram revealed 3 distinct branches consisting of (1) OspCA, OspCB, and OspCK, (2) C6-17 and 193-210A (C10), and (3) the other OspC-derived peptides (Figure 6A). This patterning reinforces the idea that the circulating antibody profiles to OspC and its immunodominant (ie, 193-210A) and subdominant linear epitopes are asynchronous and not directly correlated with each other. Moreover, there was no obvious clustering on the vertical dendrogram to indicate a relationship between antibody reactivity profiles and seropositivity categories determined in the 2-tiered Lyme disease diagnostic test (ie, IgM+/-, IgG+/-).
The HC plot of the PTLD serum samples was distinct from the diagnostic samples in that there were just 2 major horizontal branches not 3: one encompassing OspCABK and VlsE C6-17, and the other containing the OspC-derived peptides, including 193-210A (Figure 6B). This same clustering was observed when a subset of PTLD samples were evaluated by ELISA for peptide and OspC reactivity, demonstrating that the results are valid across assay platforms (Supplementary Figure 2). On the vertical dendrogram, there were 2 branches: the top branch corresponding to low peptide reactivity with or without concomitant C6-17 and OspC reactivities and the bottom branch corresponding to high peptide reactivity that were more frequently than not associated with high C6-17 reactivity. Within the bottom branch, there was a notable subbranch corresponding to 4 samples with strikingly high reactivity with 193-210A, C6-17, and most of the OspC-derived peptides (including 9-33ABK). We would tentatively classify these handful of samples as “polyreactive” [64]. In summary, HC analysis of both the diagnostic and PTLD cohorts confirms the asynchronous nature of serum antibody profiles associated with rOspC and OspC-derived peptides that may, in fact, reflect the intrinsically complicated immune response to B. burgdorferi infection in the first place.
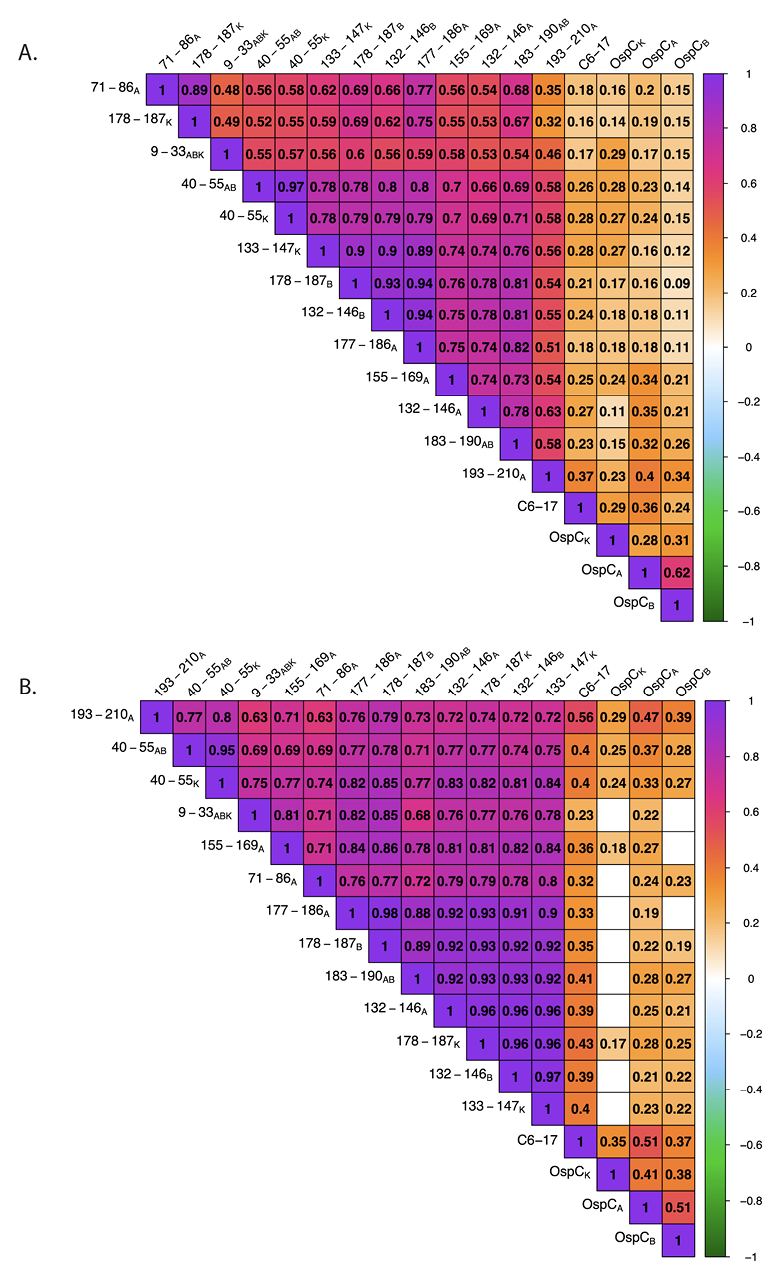
Figure 5. Correlation matrices of rOspC and OspC-derived peptide reactivity in diagnostic and PTLD serum. Diagnostic (A) and PTLD (B) serum sample MFIs (indexed and log10 transformed) were subjected to Spearman’s Rank correlation. Only significant correlation coefficients (rs) are displayed (confidence level = 0.95). The degree and direction of correlation is indicated by the color scale. Interpretations of the strength of each correlation was conducted as such: >0.7 indicates a strong correlation, 0.5 – 0.7 is moderate, 0.3 – 0.5 is weak, and 0.0 – 0.3 is negligible.
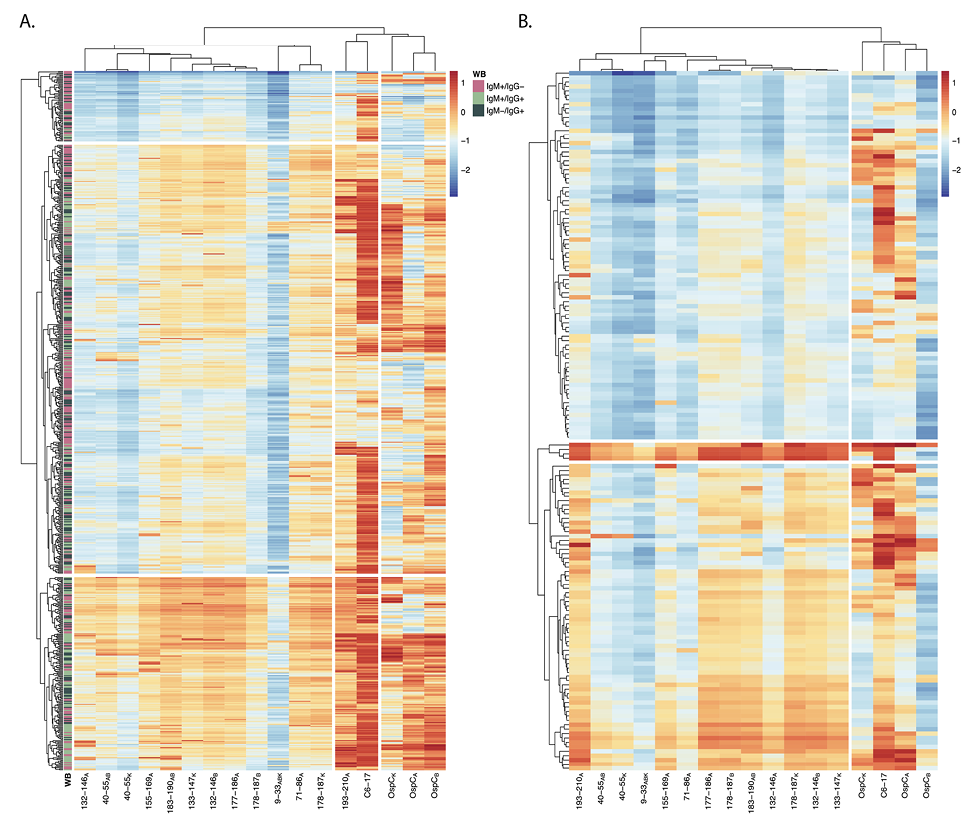
Figure 6. Hierarchical clustering of rOspC and OspC-derived reactivity in diagnostic and PTLD serum. Heatmaps were generated using diagnostic (A) and PTLD (B) serum sample MFIs (indexed and log10 transformed). Rows represent individual samples and columns represent the panel of antigens. The color scales of A and B are identical to one another (allowing for direct comparison), with 0 representing the positivity cutoff (as seen in Figure 4). Annotation of the diagnostic samples (A) with Western blot profile data (previously described in the Materials and Methods) is designated by the legend. The columns were subjected to hierarchical clustering by correlation and the rows were clustered by Euclidean distance to better visualize patterns in reactivity profiles.
Defining the breadth and the frequency of linear B-cell epitope reactivity on OspC has important implications for both Lyme disease diagnostics and vaccines. In this report, a panel of peptides representing 8 previously reported and predicted linear epitopes on OspC types A, B, and K, were examined by multiplex immunoassay for serum IgG reactivity in diagnostic (n~700) and PTLD (n~150) B. burgdorferi seropositive sample sets. The results revealed that 6 of the 8 peptides are indeed antibody targets even though high reactivity to any given peptide was confined to only a fraction (2% to 10%) of the total. The exception being peptide 193-210A, which was 26% positive (4.4-fold increase value) in the diagnostic sample set and 14% positive (3.3-fold increase value) in the PLTDs sample set. Interestingly, in the diagnostic sample set, 193-210A antibody titers did not correlate with either reactivity to OspC itself or any of the other OspC-derived peptides. In the PTLD sample set, 193-210A antibody levels did correlate with the other OspC-derived peptides but were distinct from OspC. These results have clinical implications, as they demonstrate that the human B-cell response to linear epitopes on OspC is both variable and asynchronous.
The 193-210A peptide includes the stretch of ~10 highly conserved amino acids at C-terminus of OspC, first recognized as an immunodominant epitope in neuroborreliosis patients [62]. Since then, the so-called C10 peptide and derivatives have been integrated into numerous diagnostic assays, including the Zeus VlseE1/C10 ELISA [26, 28, 29]. Dolange and colleagues recently proposed the use of high affinity C10-specific monoclonal antibodies as tools to detect circulating levels of B. burgdorferi antigens [65]. While there is little debate as to whether C10 constitutes a human B-cell epitope, the issue of whether antibodies to C10 contribute to complement-dependent, antibody-mediated killing of B. burgdorferi (“borreliacidal activity”) remains contentious. Lovrich and colleagues reported that OspC-specific borreliacidal activity (in a handful of human serum samples) was almost entirely directed against C10 [33, 34], whereas Izac and colleagues argued that, in rodents, canines, and non-human primates, C10-specific antibodies contribute little if any borreliacidal activity [66].
Although our results do not resolve this issue per se, they do raise questions about the underlying B-cell biology associated with antibodies to C10 and OspC. The fact that C10 and OspC antibody titers did not correlate with each other in the 2 serum sample sets examined suggests differences in the kinetics of antibody induction, circulating half-life, and/or plasma cell persistence. Others have suggested, at least in the case of VlsE, that there is a distinct progression of epitope reactivity over the course of B. burgdorferi infection with readily accessible epitopes targeted first and more obstructed, membrane proximal epitopes following [67]. Resolving detailed questions about the nature of the human B-cell response to OspC is becoming more feasible as single cell VH and VL repertoires are catalogued longitudinally over the course of Lyme disease [12, 68].
B-cell epitope prediction software like Discotope identifies residues 133-147 as having a high immunogenic propensity [69]. That prediction was borne out in our study, as evidenced by statistically significant reactivity of serum samples with the 3 peptides corresponding to residues 132-146AB and 133-147K. Others have reported similar findings in mice and in a few human samples. Namely, Earnhart recognized residues 136-150 (which they refer to as “loop 5”) as a target of antibodies in B. burgdorferi-infected mice [35]. In a follow-up study, the same investigators identified 2 serum samples from patients with early Lyme disease that reacted with peptides spanning 136-144 [37]. A subsequent report by Arnaboldi and colleagues did not detect binding to 3 different 15-mer OspCK-derived peptides spanning residues 131-155, possibly because their sample size was limited [28]. In our collection of almost 700 diagnostic samples, the frequency of peptide positivity was admittedly relatively low (3% to 7%). Nonetheless, antibodies targeting this epitopic region may be consequential in terms of promoting spirochete clearance [35]. For example, the potent borreliacidal mouse monoclonal antibody 16.22 recognizes OspCA residues 133-147 with the key contact points postulated to be residues 139-141 (KXK motif) [36]. Thus, a more detailed examination of B-cell epitopes within this region of OspC in human patients with Lyme disease is warranted.
It has been postulated that serum antibody profiling at the antigen and possibly even epitope level may be able to resolve different stages of Lyme disease (eg, acute, acute resolved, PTLD) [10, 67]. While the antibody profiling in our study was limited to serum IgG and IgM reactivity to OspC, OspC-derived peptides, and VlsE C6-17, the results revealed distinct patterns between the diagnostic and PTLD sample sets when subjected to HC. Most notable was the classification of the C10 peptide: in the diagnostic sample set C10 clustered with C6-17, whereas in the PTLD samples it clustered with the other OspC-derived peptides. Thus, it is tempting to speculate that the ratio of C10 to C6-17 serum IgG reactivity may have some utility in defining disease progression. By the same token, as virtually all samples tested were positive for either C10 and/or C6-17, establishing a combined C10 + C6-17 threshold value might serve to alleviate false positives in Lyme disease diagnostics based solely on serology [70, 71]. That said, we recognize that any claims to this effect are premature, considering that the serum samples employed in our study are not associated with any clinical information beyond seropositivity. Moreover, we lack a “post-Lyme healthy” cohort that Chandra and colleagues so effectively used in their study to compare with their PTLD cohort [10].
One interesting grouping that emerged from the PTLD cohort was a cluster of 4 individuals who demonstrated pan-reactivity with the OspC-derived peptides and VlsE C6-17 peptide. In follow-up analysis, those same 4 samples in question also reacted with other B. burgdorferi-derived peptides, as well as control non-B. burgdorferi-derived peptides, but not necessarily with the corresponding recombinant proteins (G. Freeman-Gallant, unpublished results). Thus, the serum antibodies from those particular individuals seemingly have a propensity for peptide antigens, possibly due to a particular bias in germline chain usage and corresponding paratopes conducive to accommodating peptides [72]. Whether or not there is a germline bias in antibodies associated with PTLD is worthy of investigation, especially in light of findings related to COVID-19. Moreover, the availability of bulk plasmablast BCR sequences, as well as single-cell paired BCR sequences from patients with Lyme disease opens the door to detailed antibody reactivity profiles and links to disease manifestations [73]. We are also exploring the possibility that these particlar samples may have profiles consistent with autoantibodies [74] or have characteristics associated with polyreactivity that might promote ligation to spirochete surface antigens [75, 76].
There are 2 notable shortcomings associated with this study that need to be underscored. First, as alluded to above, is our reliance on remnant serum samples with limited clinical attributes associated with them, rather than using samples procured as part of a prospective study of patients with Lyme disease in which serum profiles might be linked to stages of disease progression and resolution [67]. Moreover, the serum samples are biased in that they were already deemed positive in the 2-tiered Lyme disease assay, which includes VlsE and/or OspC antigens or epitopes, and from a limited geographical area (Northeast United States). Second are the intrinsic limitations associated with peptide-based epitope reactivity profiling, as detailed in a recent review [77]. Specifically, polypeptides, whether in solution or bound to a matrix, do not necessarily assume a “native” secondary structure. As a result, low or no antibody reactivity to a given peptide does not necessarily indicate that the corresponding region of the native antigen is not immunogenic. Nonetheless, we employed a variety of methods to validate our peptide epitopes wherever possible, including (1) using peptides previously reported in the literature as being reactive serum samples from patients with Lyme disease as detailed in Table 1; (2) using monoclonal antibodies (MAb) to validate the peptide epitopes [65, 78]; (3) validating epitope specificity by alanine scanning [78]; (4) coupling peptides to Luminex beads via C-terminal GGGSK-biotin linker rather than direct chemical or passive conjugation; and (5) employing commercial Lyme disease seronegative and seropositive samples as intra-assay and bead coupling confirmation controls, as described in the Materials and Methods. Despite these limitations, our study involving the profiling of >800 unique Bb+ serum samples provides important insights into epitope usage across the surface of OspC that will have implications for diagnostic and vaccine implementation.
We thank Dr. William Lee and other members of the Diagnostic Immunology Laboratory at the Wadsworth Center for providing residual diagnostic serum samples. We gratefully acknowledge Dr. John Martignetti and Lisa Arrigo of the Nuvance Health Lyme disease Biobank for generously providing PTLD serum samples. We thank Elizabeth Cavosie (Wadsworth Center) for administrative assistance and Greta Van Slyke (Wadsworth Center) for establishing Luminex protocols.
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, Contracts 75N93019C00040 and 75N93024C00069. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors have no conflicts to declare.
Supplementary materials are available at the Pathogens and Immunity website. Supplementary data may be provided by the authors to benefit the reader. Supplementary data are not copyedited and are the sole responsibility of the authors. Questions or comments related to supplementary materials should be addressed to the corresponding author.
Supplementary Figures and Tables
1. Kugeler KJ, Schwartz AM, Delorey MJ, Mead PS, Hinckley AF. Estimating the Frequency of Lyme Disease Diagnoses, United States, 2010-2018. Emerg Infect Dis. 2021;27(2):616-9. doi: 10.3201/eid2702.202731. PubMed PMID: 33496229; PMCID: PMC7853543.
2. Bobe JR, Jutras BL, Horn EJ, Embers ME, Bailey A, Moritz RL, Zhang Y, Soloski MJ, Ostfeld RS, Marconi RT, Aucott J, Ma’ayan A, Keesing F, Lewis K, Ben Mamoun C, Rebman AW, McClune ME, Breitschwerdt EB, Reddy PJ, Maggi R, Yang F, Nemser B, Ozcan A, Garner O, Di Carlo D, Ballard Z, Joung HA, Garcia-Romeu A, Griffiths RR, Baumgarth N, Fallon BA. Recent Progress in Lyme Disease and Remaining Challenges. Front Med (Lausanne). 2021;8:666554. doi: 10.3389/fmed.2021.666554. PubMed PMID: 34485323; PMCID: PMC8416313.
3. Radolf JD, Strle K, Lemieux JE, Strle F. Lyme Disease in Humans. Curr Issues Mol Biol. 2021;42:333-84. doi: 10.21775/cimb.042.333. PubMed PMID: 33303701; PMCID: PMC7946767.
4. Steere AC, Strle F, Wormser GP, Hu LT, Branda JA, Hovius JW, Li X, Mead PS. Lyme borreliosis. Nat Rev Dis Primers. 2016;2:16090. doi: 10.1038/nrdp.2016.90. PubMed PMID: 27976670; PMCID: PMC5539539.
5. Lochhead RB, Strle K, Arvikar SL, Weis JJ, Steere AC. Lyme arthritis: linking infection, inflammation and autoimmunity. Nat Rev Rheumatol. 2021;17(8):449-61. doi: 10.1038/s41584-021-00648-5. PubMed PMID: 34226730; PMCID: PMC9488587.
6. Klempner MS, Hu LT, Evans J, Schmid CH, Johnson GM, Trevino RP, Norton D, Levy L, Wall D, McCall J, Kosinski M, Weinstein A. Two controlled trials of antibiotic treatment in patients with persistent symptoms and a history of Lyme disease. N Engl J Med. 2001;345(2):85-92. doi: 10.1056/NEJM200107123450202. PubMed PMID: 11450676.
7. Marques A. Chronic Lyme disease: a review. Infect Dis Clin North Am. 2008;22(2):341-60, vii-viii. doi: 10.1016/j.idc.2007.12.011. PubMed PMID: 18452806; PMCID: PMC2430045.
8. Aucott JN, Crowder LA, Kortte KB. Development of a foundation for a case definition of post-treatment Lyme disease syndrome. Int J Infect Dis. 2013;17(6):e443-9. doi: 10.1016/j.ijid.2013.01.008. PubMed PMID: 23462300.
9. Aucott JN, Yang T, Yoon I, Powell D, Geller SA, Rebman AW. Risk of post-treatment Lyme disease in patients with ideally-treated early Lyme disease: A prospective cohort study. Int J Infect Dis. 2022;116:230-7. doi: 10.1016/j.ijid.2022.01.033. PubMed PMID: 35066160.
10. Chandra A, Wormser GP, Marques AR, Latov N, Alaedini A. Anti-Borrelia burgdorferi antibody profile in post-Lyme disease syndrome. Clin Vaccine Immunol. 2011;18(5):767-71. doi: 10.1128/CVI.00002-11. PubMed PMID: 21411605; PMCID: PMC3122515.
11. Ghosh R, Joung HA, Goncharov A, Palanisamy B, Ngo K, Pejcinovic K, Krockenberger N, Horn EJ, Garner OB, Ghazal E, O’Kula A, Arnaboldi PM, Dattwyler RJ, Ozcan A, Di Carlo D. Rapid single-tier serodiagnosis of Lyme disease. Nat Commun. 2024;15(1):7124. doi: 10.1038/s41467-024-51067-5. PubMed PMID: 39164226; PMCID: PMC11336255.
12. Blum LK, Adamska JZ, Martin DS, Rebman AW, Elliott SE, Cao RRL, Embers ME, Aucott JN, Soloski MJ, Robinson WH. Robust B Cell Responses Predict Rapid Resolution of Lyme Disease. Front Immunol. 2018;9:1634. doi: 10.3389/fimmu.2018.01634. PubMed PMID: 30072990; PMCID: PMC6060717.
13. Bockenstedt LK, Wooten RM, Baumgarth N. Immune Response to Borrelia: Lessons from Lyme Disease Spirochetes. Curr Issues Mol Biol. 2021;42:145-90. doi: 10.21775/cimb.042.145. PubMed PMID: 33289684; PMCID: PMC10842262.
14. Embers ME, Hasenkampf NR, Jacobs MB, Philipp MT. Dynamic longitudinal antibody responses during Borrelia burgdorferi infection and antibiotic treatment of rhesus macaques. Clin Vaccine Immunol. 2012;19(8):1218-26. doi: 10.1128/CVI.00228-12. PubMed PMID: 22718128; PMCID: PMC3416093.
15. Jiang R, Meng H, Raddassi K, Fleming I, Hoehn KB, Dardick KR, Belperron AA, Montgomery RR, Shalek AK, Hafler DA, Kleinstein SH, Bockenstedt LK. Single-cell immunophenotyping of the skin lesion erythema migrans identifies IgM memory B cells. JCI Insight. 2021;6(12). doi: 10.1172/jci.insight.148035. PubMed PMID: 34061047; PMCID: PMC8262471.
16. Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun. 2008;76(8):3374-89. doi: 10.1128/IAI.00048-08. PubMed PMID: 18474646; PMCID: PMC2493225.
17. Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Vanraden MJ, Stewart P, Rosa P. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun. 2006;74(6):3554-64. doi: 10.1128/IAI.01950-05. PubMed PMID: 16714588; PMCID: PMC1479285.
18. Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113(2):220-30. doi: 10.1172/JCI19894. PubMed PMID: 14722614; PMCID: PMC311436.
19. Tilly K, Bestor A, Jewett MW, Rosa P. Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect Immun. 2007;75(3):1517-9. doi: 10.1128/IAI.01725-06. PubMed PMID: 17158906; PMCID: PMC1828573.
20. Earnhart CG, Leblanc DV, Alix KE, Desrosiers DC, Radolf JD, Marconi RT. Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Mol Microbiol. 2010;76(2):393-408. doi: 10.1111/j.1365-2958.2010.07103.x. PubMed PMID: 20199597; PMCID: PMC2917209.
21. Caine JA, Coburn J. Multifunctional and Redundant Roles of Borrelia burgdorferi Outer Surface Proteins in Tissue Adhesion, Colonization, and Complement Evasion. Front Immunol. 2016;7:442. doi: 10.3389/fimmu.2016.00442. PubMed PMID: 27818662; PMCID: PMC5073149.
22. Caine JA, Lin YP, Kessler JR, Sato H, Leong JM, Coburn J. Borrelia burgdorferi outer surface protein C (OspC) binds complement component C4b and confers bloodstream survival. Cell Microbiol. 2017;19(12). doi: 10.1111/cmi.12786. PubMed PMID: 28873507; PMCID: PMC5680108.
23. Lin YP, Tan X, Caine JA, Castellanos M, Chaconas G, Coburn J, Leong JM. Strain-specific joint invasion and colonization by Lyme disease spirochetes is promoted by outer surface protein C. PLoS Pathog. 2020;16(5):e1008516. doi: 10.1371/journal.ppat.1008516. PubMed PMID: 32413091; PMCID: PMC7255614.
24. Tan X, Castellanos M, Chaconas G. Choreography of Lyme Disease Spirochete Adhesins To Promote Vascular Escape. Microbiol Spectr. 2023;11(4):e0125423. doi: 10.1128/spectrum.01254-23. PubMed PMID: 37255427; PMCID: PMC10434219.
25. Branda JA, Steere AC. Laboratory Diagnosis of Lyme Borreliosis. Clin Microbiol Rev. 2021;34(2). doi: 10.1128/CMR.00018-19. PubMed PMID: 33504503; PMCID: PMC7849240.
26. Hahm JB, Breneman JWt, Liu J, Rabkina S, Zheng W, Zhou S, Walker RP, Kaul R. A Fully Automated Multiplex Assay for Diagnosis of Lyme Disease with High Specificity and Improved Early Sensitivity. J Clin Microbiol. 2020;58(5). doi: 10.1128/JCM.01785-19. PubMed PMID: 32132190; PMCID: PMC7180237.
27. Bacon RM, Biggerstaff BJ, Schriefer ME, Gilmore RD, Jr., Philipp MT, Steere AC, Wormser GP, Marques AR, Johnson BJ. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J Infect Dis. 2003;187(8):1187-99. doi: 10.1086/374395. PubMed PMID: 12695997; PMCID: PMC7109709.
28. Arnaboldi PM, Seedarnee R, Sambir M, Callister SM, Imparato JA, Dattwyler RJ. Outer surface protein C peptide derived from Borrelia burgdorferi sensu stricto as a target for serodiagnosis of early lyme disease. Clin Vaccine Immunol. 2013;20(4):474-81. doi: 10.1128/CVI.00608-12. PubMed PMID: 23365204; PMCID: PMC3623407.
29. Baarsma ME, Vrijlandt A, Ursinus J, Zaaijer HL, Jurriaans S, van Dam AP, Hovius JW. Diagnostic performance of the ZEUS Borrelia VlsE1/pepC10 assay in European LB patients: a case-control study. Eur J Clin Microbiol Infect Dis. 2022;41(3):387-93. doi: 10.1007/s10096-021-04372-6. PubMed PMID: 34806121.
30. Coleman AS, Rossmann E, Yang X, Song H, Lamichhane CM, Iyer R, Schwartz I, Pal U. BBK07 immunodominant peptides as serodiagnostic markers of Lyme disease. Clin Vaccine Immunol. 2011;18(3):406-13. doi: 10.1128/CVI.00461-10. PubMed PMID: 21177911; PMCID: PMC3067378.
31. Mathiesen MJ, Christiansen M, Hansen K, Holm A, Asbrink E, Theisen M. Peptide-based OspC enzyme-linked immunosorbent assay for serodiagnosis of Lyme borreliosis. J Clin Microbiol. 1998;36(12):3474-9. doi: 10.1128/JCM.36.12.3474-3479.1998. PubMed PMID: 9817857; PMCID: PMC105224.
32. O’Bier NS, Hatke AL, Camire AC, Marconi RT. Human and Veterinary Vaccines for Lyme Disease. Curr Issues Mol Biol. 2021;42:191-222. doi: 10.21775/cimb.042.191. PubMed PMID: 33289681; PMCID: PMC7946718.
33. Jobe DA, Lovrich SD, Schell RF, Callister SM. C-terminal region of outer surface protein C binds borreliacidal antibodies in sera from patients with Lyme disease. Clin Diagn Lab Immunol. 2003;10(4):573-8. doi: 10.1128/cdli.10.4.573-578.2003. PubMed PMID: 12853388; PMCID: PMC164245.
34. Lovrich SD, Jobe DA, Schell RF, Callister SM. Borreliacidal OspC antibodies specific for a highly conserved epitope are immunodominant in human lyme disease and do not occur in mice or hamsters. Clin Diagn Lab Immunol. 2005;12(6):746-51. doi: 10.1128/CDLI.12.6.746-751.2005. PubMed PMID: 15939749; PMCID: PMC1151971.
35. Earnhart CG, Buckles EL, Dumler JS, Marconi RT. Demonstration of OspC type diversity in invasive human lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC murine antibody response. Infect Immun. 2005;73(12):7869-77. doi: 10.1128/IAI.73.12.7869-7877.2005. PubMed PMID: 16299277; PMCID: PMC1307023.
36. Yang X, Li Y, Dunn JJ, Luft BJ. Characterization of a unique borreliacidal epitope on the outer surface protein C of Borrelia burgdorferi. FEMS Immunol Med Microbiol. 2006;48(1):64-74. doi: 10.1111/j.1574-695X.2006.00122.x. PubMed PMID: 16965353.
37. Buckles EL, Earnhart CG, Marconi RT. Analysis of antibody response in humans to the type A OspC loop 5 domain and assessment of the potential utility of the loop 5 epitope in Lyme disease vaccine development. Clin Vaccine Immunol. 2006;13(10):1162-5. doi: 10.1128/CVI.00099-06. PubMed PMID: 17028218; PMCID: PMC1595320.
38. Earnhart CG, Marconi RT. Construction and analysis of variants of a polyvalent Lyme disease vaccine: approaches for improving the immune response to chimeric vaccinogens. Vaccine. 2007;25(17):3419-27. doi: 10.1016/j.vaccine.2006.12.051. PubMed PMID: 17239505; PMCID: PMC2696934.
39. Pulzova L, Flachbartova Z, Bencurova E, Potocnakova L, Comor L, Schreterova E, Bhide M. Identification of B-cell epitopes of Borrelia burgdorferi outer surface protein C by screening a phage-displayed gene fragment library. Microbiol Immunol. 2016;60(10):669-77. doi: 10.1111/1348-0421.12438. PubMed PMID: 27619624.
40. Norek A, Janda L. Epitope mapping of Borrelia burgdorferi OspC protein in homodimeric fold. Protein Sci. 2017;26(4):796-806. doi: 10.1002/pro.3125. PubMed PMID: 28142214; PMCID: PMC5368064.
41. Izac JR, Oliver LD, Jr., Earnhart CG, Marconi RT. Identification of a defined linear epitope in the OspA protein of the Lyme disease spirochetes that elicits bactericidal antibody responses: Implications for vaccine development. Vaccine. 2017;35(24):3178-85. doi: 10.1016/j.vaccine.2017.04.079. PubMed PMID: 28479174; PMCID: PMC8203411.
42. Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Schwab E, Will G, Wanner G. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61(5):2182-91. doi: 10.1128/iai.61.5.2182-2191.1993. PubMed PMID: 8478108; PMCID: PMC280819.
43. Barbour AG, Travinsky B. Evolution and distribution of the ospC Gene, a transferable serotype determinant of Borrelia burgdorferi. mBio. 2010;1(4). doi: 10.1128/mBio.00153-10. PubMed PMID: 20877579; PMCID: PMC2945197.
44. Cerar T, Strle F, Stupica D, Ruzic-Sabljic E, McHugh G, Steere AC, Strle K. Differences in Genotype, Clinical Features, and Inflammatory Potential of Borrelia burgdorferi sensu stricto Strains from Europe and the United States. Emerg Infect Dis. 2016;22(5):818-27. doi: 10.3201/eid2205.151806. PubMed PMID: 27088349; PMCID: PMC4861522.
45. Seinost G, Dykhuizen DE, Dattwyler RJ, Golde WT, Dunn JJ, Wang IN, Wormser GP, Schriefer ME, Luft BJ. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun. 1999;67(7):3518-24. doi: 10.1128/IAI.67.7.3518-3524.1999. PubMed PMID: 10377134; PMCID: PMC116539.
46. Hanincova K, Mukherjee P, Ogden NH, Margos G, Wormser GP, Reed KD, Meece JK, Vandermause MF, Schwartz I. Multilocus sequence typing of Borrelia burgdorferi suggests existence of lineages with differential pathogenic properties in humans. PLoS One. 2013;8(9):e73066. doi: 10.1371/journal.pone.0073066. PubMed PMID: 24069170; PMCID: PMC3775742.
47. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390(6660):580-6. doi: 10.1038/37551. PubMed PMID: 9403685.
48. Anderson JF, Flavell RA, Magnarelli LA, Barthold SW, Kantor FS, Wallich R, Persing DH, Mathiesen D, Fikrig E. Novel Borrelia burgdorferi isolates from Ixodes scapularis and Ixodes dentatus ticks feeding on humans. J Clin Microbiol. 1996;34(3):524-9. doi: 10.1128/jcm.34.3.524-529.1996. PubMed PMID: 8904407; PMCID: PMC228839.
49. Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308(13):733-40. doi: 10.1056/NEJM198303313081301. PubMed PMID: 6828118.
50. Rudolph MJ, Davis SA, Haque HME, Weis DD, Vance DJ, Piazza CL, Ejemel M, Cavacini L, Wang Y, Mbow ML, Gilmore RD, Mantis NJ. Structural Elucidation of a Protective B Cell Epitope on Outer Surface Protein C (OspC) of the Lyme Disease Spirochete, Borreliella burgdorferi. mBio. 2023;14(2):e0298122. doi: 10.1128/mbio.02981-22. PubMed PMID: 36976016; PMCID: PMC10128040.
51. Kringelum JV, Lundegaard C, Lund O, Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS computational biology. 2012;8(12):e1002829. doi: 10.1371/journal.pcbi.1002829. PubMed PMID: 23300419; PMCID: PMC3531324.
52. Gomes-Solecki MJ, Meirelles L, Glass J, Dattwyler RJ. Epitope length, genospecies dependency, and serum panel effect in the IR6 enzyme-linked immunosorbent assay for detection of antibodies to Borrelia burgdorferi. Clin Vaccine Immunol. 2007;14(7):875-9. doi: 10.1128/CVI.00122-07. PubMed PMID: 17538122; PMCID: PMC1951069.
53. Nemeth KL, Yauney E, Rock JM, Bievenue R, Parker MM, Styer LM. Use of Self-Collected Dried Blood Spots and a Multiplex Microsphere Immunoassay to Measure IgG Antibody Response to COVID-19 Vaccines. Microbiol Spectr. 2023;11(1):e0133622. doi: 10.1128/spectrum.01336-22. PubMed PMID: 36622204; PMCID: PMC9927373.
54. Yates JL, Ehrbar DJ, Hunt DT, Girardin RC, Dupuis AP, 2nd, Payne AF, Sowizral M, Varney S, Kulas KE, Demarest VL, Howard KM, Carson K, Hales M, Ejemel M, Li Q, Wang Y, Peredo-Wende R, Ramani A, Singh G, Strle K, Mantis NJ, McDonough KA, Lee WT. Serological analysis reveals an imbalanced IgG subclass composition associated with COVID-19 disease severity. Cell Rep Med. 2021;2(7):100329. doi: 10.1016/j.xcrm.2021.100329. PubMed PMID: 34151306; PMCID: PMC8205277.
55. R core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing,. 2023.
56. Wickham H, Bryan J. readxl: Read Excel Files 1.4.3. 2023. https://readxl.tidyverse.org, https://github.com/tidyverse/readxl.
57. Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T, Miller E, Bache S, Müller K, Ooms J, Robinson D, Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. Welcome to the Tidyverse. Journal of Open Source Software. 2019;4(43):1686. doi: 10.21105/joss.01686.
58. Wei T, Simko V, Levy M, Xie Y, Jin Y, Zemla J. Package ‘corrplot’. Statistician. 2017;56(316):e24.
59. Kolde R. pheatmap: Pretty Heatmaps 2018 [updated 2018-05-19]. Available from: https://cran.r-project.org/web/packages/pheatmap/index.html.
60. Lemieux JE, Huang W, Hill N, Cerar T, Freimark L, Hernandez S, Luban M, Maraspin V, Bogovic P, Ogrinc K, Ruzic-Sabljic E, Lapierre P, Lasek-Nesselquist E, Singh N, Iyer R, Liveris D, Reed KD, Leong JM, Branda JA, Steere AC, Wormser GP, Strle F, Sabeti PC, Schwartz I, Strle K. Whole genome sequencing of human Borrelia burgdorferi isolates reveals linked blocks of accessory genome elements located on plasmids and associated with human dissemination. PLoS Pathog. 2023;19(8):e1011243. doi: 10.1371/journal.ppat.1011243. PubMed PMID: 37651316; PMCID: PMC10470944.
61. Wormser GP, Brisson D, Liveris D, Hanincova K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis. 2008;198(9):1358-64. doi: 10.1086/592279. PubMed PMID: 18781866; PMCID: PMC2776734.
62. Mathiesen MJ, Holm A, Christiansen M, Blom J, Hansen K, Ostergaard S, Theisen M. The dominant epitope of Borrelia garinii outer surface protein C recognized by sera from patients with neuroborreliosis has a surface-exposed conserved structural motif. Infect Immun. 1998;66(9):4073-9. doi: 10.1128/IAI.66.9.4073-4079.1998. PubMed PMID: 9712750; PMCID: PMC108488.
63. Earnhart CG, Rhodes DV, Smith AA, Yang X, Tegels B, Carlyon JA, Pal U, Marconi RT. Assessment of the potential contribution of the highly conserved C-terminal motif (C10) of Borrelia burgdorferi outer surface protein C in transmission and infectivity. Pathog Dis. 2014;70(2):176-84. doi: 10.1111/2049-632X.12119. PubMed PMID: 24376161; PMCID: PMC4497580.
64. Dimitrov JD, Planchais C, Roumenina LT, Vassilev TL, Kaveri SV, Lacroix-Desmazes S. Antibody polyreactivity in health and disease: statu variabilis. J Immunol. 2013;191(3):993-9. doi: 10.4049/jimmunol.1300880. PubMed PMID: 23873158.
65. Dolange V, Simon S, Morel N. Detection of Borrelia burgdorferi antigens in tissues and plasma during early infection in a mouse model. Sci Rep. 2021;11(1):17368. doi: 10.1038/s41598-021-96861-z. PubMed PMID: 34462491; PMCID: PMC8405660.
66. Izac JR, Camire AC, Earnhart CG, Embers ME, Funk RA, Breitschwerdt EB, Marconi RT. Analysis of the antigenic determinants of the OspC protein of the Lyme disease spirochetes: Evidence that the C10 motif is not immunodominant or required to elicit bactericidal antibody responses. Vaccine. 2019;37(17):2401-7. doi: 10.1016/j.vaccine.2019.02.007. PubMed PMID: 30922701; PMCID: PMC6453540.
67. Jacek E, Tang KS, Komorowski L, Ajamian M, Probst C, Stevenson B, Wormser GP, Marques AR, Alaedini A. Epitope-Specific Evolution of Human B Cell Responses to Borrelia burgdorferi VlsE Protein from Early to Late Stages of Lyme Disease. J Immunol. 2016;196(3):1036-43. doi: 10.4049/jimmunol.1501861. PubMed PMID: 26718339; PMCID: PMC4722499.
68. Kirpach J, Colone A, Burckert JP, Faison WJ, Dubois A, Sinner R, Reye AL, Muller CP. Detection of a Low Level and Heterogeneous B Cell Immune Response in Peripheral Blood of Acute Borreliosis Patients With High Throughput Sequencing. Front Immunol. 2019;10:1105. doi: 10.3389/fimmu.2019.01105. PubMed PMID: 31156648; PMCID: PMC6532064.
69. Hoie MH, Gade FS, Johansen JM, Wurtzen C, Winther O, Nielsen M, Marcatili P. DiscoTope-3.0: improved B-cell epitope prediction using inverse folding latent representations. Front Immunol. 2024;15:1322712. doi: 10.3389/fimmu.2024.1322712. PubMed PMID: 38390326; PMCID: PMC10882062.
70. Rebman AW, Crowder LA, Kirkpatrick A, Aucott JN. Characteristics of seroconversion and implications for diagnosis of post-treatment Lyme disease syndrome: acute and convalescent serology among a prospective cohort of early Lyme disease patients. Clin Rheumatol. 2015;34(3):585-9. doi: 10.1007/s10067-014-2706-z. PubMed PMID: 24924604.
71. Reifert J, Kamath K, Bozekowski J, Lis E, Horn EJ, Granger D, Theel ES, Shon J, Sawyer JR, Daugherty PS. Serum Epitope Repertoire Analysis Enables Early Detection of Lyme Disease with Improved Sensitivity in an Expandable Multiplex Format. J Clin Microbiol. 2021;59(2). doi: 10.1128/JCM.01836-20. PubMed PMID: 33148704; PMCID: PMC8111119.
72. Cobaugh CW, Almagro JC, Pogson M, Iverson B, Georgiou G. Synthetic antibody libraries focused towards peptide ligands. J Mol Biol. 2008;378(3):622-33. doi: 10.1016/j.jmb.2008.02.037. PubMed PMID: 18384812; PMCID: PMC2494707.
73. Blum JS, Fiani ML, Stahl PD. Proteolytic cleavage of ricin A chain in endosomal vesicles. Evidence for the action of endosomal proteases at both neutral and acidic pH. J Biol Chem. 1991;266(33):22091-5. PubMed PMID: 1939230.
74. Trier NH, Houen G. Antibody Cross-Reactivity in Auto-Immune Diseases. Int J Mol Sci. 2023;24(17). doi: 10.3390/ijms241713609. PubMed PMID: 37686415; PMCID: PMC10487534.
75. Boughter CT, Borowska MT, Guthmiller JJ, Bendelac A, Wilson PC, Roux B, Adams EJ. Biochemical patterns of antibody polyreactivity revealed through a bioinformatics-based analysis of CDR loops. Elife. 2020;9. doi: 10.7554/eLife.61393. PubMed PMID: 33169668; PMCID: PMC7755423.
76. Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, Nussenzweig MC. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467(7315):591-5. doi: 10.1038/nature09385. PubMed PMID: 20882016; PMCID: PMC3699875.
77. Iaculli D, Ballet S. Peptide libraries: from epitope mapping to in-depth high-throughput analysis. Trends Pharmacol Sci. 2024;45(7):579-82. doi: 10.1016/j.tips.2024.04.004. PubMed PMID: 38724411.
78. Vance DJ, Freeman-Gallant G, McCarthy K, Piazza CL, Chen Y, Vorauer C, Muriuki B, Rudolph MJ, Cavacini L, Guttman M, Mantis NJ. 2024. doi: 10.1101/2024.11.06.622094.
79. Ikushima M, Yamada F, Kawahashi S, Okuyama Y, Matsui K. Antibody response to OspC-I synthetic peptide derived from outer surface protein C of Borrelia burgdorferi in sera from Japanese forestry workers. Epidemiol Infect. 1999;122(3):429-33. doi: 10.1017/s0950268899002320. PubMed PMID: 10459646; PMCID: PMC2809637.
80. Tokarz R, Mishra N, Tagliafierro T, Sameroff S, Caciula A, Chauhan L, Patel J, Sullivan E, Gucwa A, Fallon B, Golightly M, Molins C, Schriefer M, Marques A, Briese T, Lipkin WI. A multiplex serologic platform for diagnosis of tick-borne diseases. Sci Rep. 2018;8(1):3158. doi: 10.1038/s41598-018-21349-2. PubMed PMID: 29453420; PMCID: PMC5816631.
81. Yu Z, Carter JM, Sigal LH, Stein S. Multi-well ELISA based on independent peptide antigens for antibody capture. Application to Lyme disease serodiagnosis. J Immunol Methods. 1996;198(1):25-33. doi: 10.1016/0022-1759(96)00140-8. PubMed PMID: 8914594.
82. Baum E, Randall AZ, Zeller M, Barbour AG. Inferring epitopes of a polymorphic antigen amidst broadly cross-reactive antibodies using protein microarrays: a study of OspC proteins of Borrelia burgdorferi. PLoS One. 2013;8(6):e67445. doi: 10.1371/journal.pone.0067445. PubMed PMID: 23826301; PMCID: PMC3691210.
Submitted September 7, 2024 | Accepted January 7, 2025 | Published February 14, 2025
Copyright © 2025 The Authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License.