Marc Antoine Jean Juste1*, Yvetot Joseph1*, Dominique Lespinasse1, Alexandra Apollon1, Parmida Jamshidi2, Myung Hee Lee2, Maureen Ward2, Esther Brill2, Yanique Duffus2, Uche Chukwukere2, Ali Danesh2, Winiffer Conce Alberto2, Daniel W. Fitzgerald2, Jean W. Pape1,2, R. Brad Jones2,
Kathryn Dupnik2
1 GHESKIO Centers, Port au Prince, Haiti
2 Department of Medicine, Weill Cornell Medicine, New York, NY
*There are 2 co-first authors. Dr. Jean Juste is listed first because the identification cohort for which he supervised enrollment contributed a larger number of participants to this study.
Kathryn M. Dupnik
kad9040@med.cornell.edu
Juste MAJ, Joseph Y, Lespinasse D, Apollon A, Jamshidi P, Lee MH, Ward M, Brill E, Duffus Y, Chukwukere U, Danesh A, Alberto WC, Fitzgerald DW, Pape JW, Jones RB, Dupnik K. People Living With HIV Have More Intact HIV DNA in Circulating CD4+ T Cells if They Have History of Pulmonary Tuberculosis. Pathogens and Immunity. 2024;9(2):172–193. doi: 10.20411/pai.v9i2.722
10.20411/pai.v9i2.722
Background: A primary barrier to curing HIV is the HIV reservoir. The leading infectious cause of death worldwide for people living with HIV is tuberculosis (TB), but we do not know how TB impacts the HIV reservoir.
Methods: Participants in identification and validation cohorts were selected from previously enrolled studies at Groupe Haïtien d’Étude du Sarcome de Kaposi et des Infections Opportunistes (GHESKIO) in Port au Prince, Haiti. Intact and non-intact proviral DNA were quantified using droplet digital PCR of peripheral blood mononuclear cell (PBMC)-derived CD4+ T cells.
Kruskal-Wallis tests were used to compare medians with tobit regression for censoring.
Results: In the identification cohort, we found that people living with HIV with a history of active pulmonary TB (n=19) had higher levels of intact provirus than people living with HIV without a history of active TB (n=47) (median 762; IQR, 183-1173 vs 117; IQR, 24-279 intact provirus per million CD4, respectively; P=0.0001). This difference also was seen in the validation cohort (n=31), (median 102; IQR, 0-737 vs 0; IQR, 0-24.5 intact provirus per million CD4, P=0.03) for TB vs no-TB history groups, respectively. The frequencies of CD4+ T cells with any detectable proviral fragment was directly proportional to the levels of interleukin-1 beta (r=0.524, P= 0.0025) and interleukin-2 (r=0.622, P=0.0002).
Conclusions: People living with HIV with a history of active pulmonary TB have more HIV provirus in their circulating CD4+ T cells, even years after TB cure. We need to characterize which CD4+ T cells are harboring intact provirus to consider the impact of T cell-targeting HIV cure interventions for people living in TB-endemic areas.
HIV; tuberculosis; virus latency; proviruses; Haiti; interleukin-1; interleukin-2
The primary barrier to curing HIV infection is the pool of intact HIV proviruses integrated into host cell DNA throughout the bodies of people living with HIV, called the HIV reservoir. Larger HIV reservoirs correlate with adverse HIV-related disease outcomes [1–4]. HIV reservoir size is related to peak viral load, duration of time with HIV infection, and time living with HIV prior to starting antiretroviral therapy (ART) [5]. Reservoir size has also been shown to be inversely correlated to the nadir CD4 count [6], likely a proxy for longer duration of infection. Recent studies show that timing of initiation of combination ART (cART) (early vs late) can impact not only the HIV reservoir quantity [7] but also the qualities of integrated proviral DNA — including the fraction that is intact versus fragmented [8].
Studies have compared changes in the reservoir size in response to antigen stimulation such as vaccination [9, 10], and we have data on the impact of a limited number of coinfections on the HIV reservoir. For example, cytomegalovirus and Epstein-Barr virus viremia have been associated with a decrease in the rate of decay of the HIV reservoir in men [11, 12]. The leading infectious cause of death worldwide for people living with HIV is tuberculosis (TB) [13]. One group used quantitative PCR of total HIV DNA in peripheral blood mononuclear cells (PBMC) to approximate reservoir size [3] in people in Uganda with and without active pulmonary TB and did not see a statistically significant difference [14]. However, in a cohort in China, people living with HIV with a history of TB had a higher level of circulating HIV DNA in PBMC than people without TB, but they also had a higher viral load pre-ART [15]. The association of increased HIV viral load and active TB has been described [16, 17] and is a potential confounder of the latter reported association of TB with reservoir size.
There are multiple assays to measure the HIV reservoir in CD4+ T cells [18], but the emerging gold standard is the intact proviral DNA assay (IPDA), which distinguishes intact from defective proviruses [19, 20]. We used a modified IPDA [21] to measure intact and fragment HIV provirus in circulating CD4+ T cells of people living with HIV in a resource-limited setting with a TB syndemic. Our objective was to determine how quantities of CD4+ T cells containing HIV provirus vary in people with a history of active bacteriologically confirmed pulmonary TB disease concurrent with or after diagnosis of HIV. Because of the overall worse outcomes for people living with HIV who have a history of TB [22], we hypothesized that proviral load would be higher in people living with HIV with a history of TB.
The prevalence of HIV in adults in Haiti was 1.7% in 2023 [23]. Haiti has the highest rate of TB in the Americas. In 2022, there were 154 new cases of TB per 100,000 people, and 15% of those individuals were living with HIV [24]. The Haitian Group for the Study of Kaposi’s Sarcoma and Opportunistic Infections (with the French acronym of GHESKIO for Groupe Haïtien d’Étude du Sarcome de Kaposi et des Infections Opportunistes) in Port au Prince, Haiti, is a Haitian non-governmental organization that was founded in 1982 and is the largest provider of HIV and TB care in the Americas [25].
This study was approved by institutional review boards at Weill Cornell Medicine (protocols 1401014658 and 1612017803) and GHESKIO. All study participants provided written informed consent [26]. There was no difference in clinical care between participants who did or did not decide to participate in either of these studies. All participants were offered ART and anti-TB treatment and continuing care for HIV and TB per the Haitian national guidelines and the GHESKIO standard of care at the time, which adhered to World Health Organization recommendations. All people at GHESKIO who are diagnosed with HIV and do not have active TB are prescribed preventive therapy for treatment of presumptive latent TB because of the high rate of TB in the community, and prior studies at GHESKIO that have documented the survival benefit of universal TB preventive treatment for people living with HIV. [27]
For the identification cohort, we designed a nested case-control study from a parent cohort study of TB called the Tri-institutional Tuberculosis Research Unit (TBRU) [28–30] (Supplementary Figure 1, Supplemental Methods). TB was defined as pulmonary TB that had been bacteriologically confirmed by positive sputum TB GeneXpert, positive sputum smear for acid-fast bacilli, or sputum culture that grew Mycobacterium tuberculosis. Latent TB testing was performed with the Quantiferon-TB Gold In-Tube interferon-gamma release assay (IGRA) (Qiagen). Blood was collected using heparin as an anticoagulant for the isolation of plasma and PBMCs.
Participants for the case-control study were recruited between June 2015 and January 2019 during which time their PBMCs were collected and cryopreserved.

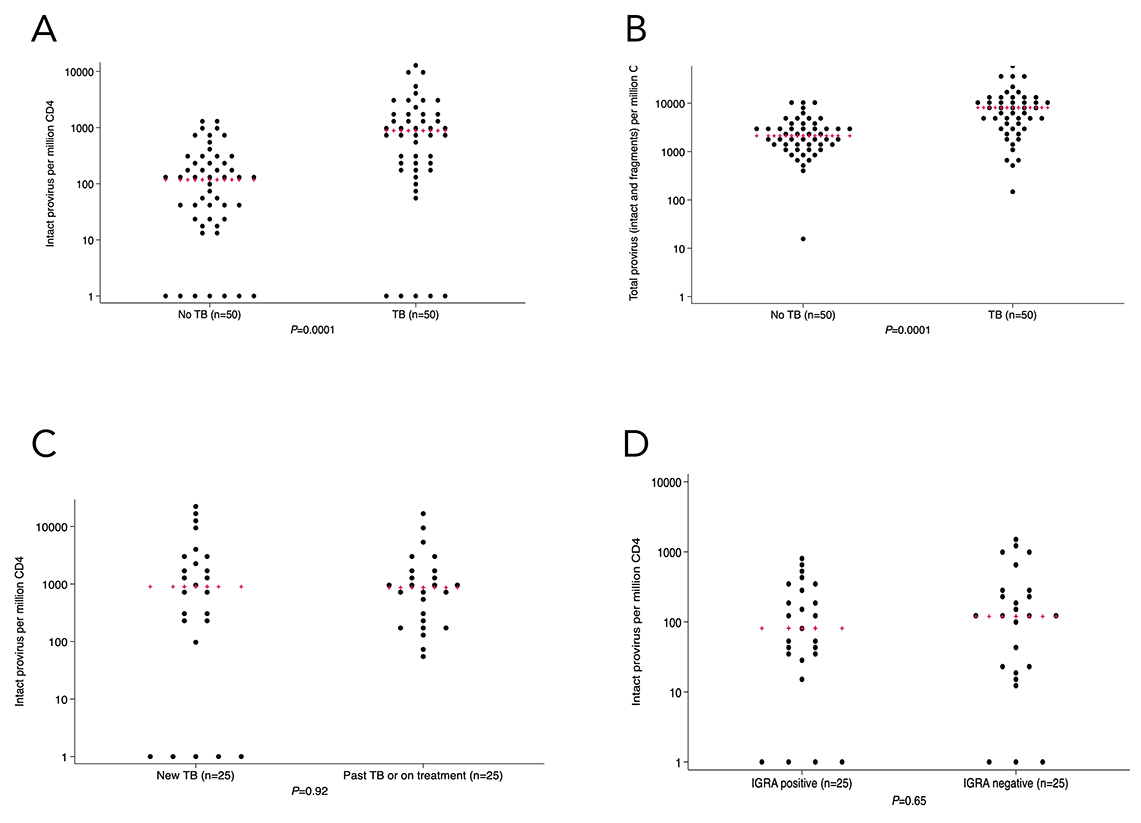
Figure 1. In the identification cohort (n=100) there was a larger number of circulating CD4+ T cells with intact (1A) and total (1B) provirus in people living with HIV with TB or TB history. There was no statistically significant difference in intact provirus levels when comparing people with active vs history of TB (1C). There was also no statistically significant difference in intact provirus levels between people with no history of TB with reactive vs non-reactive TB interferon-gamma release assay (1D). Kruskal-Wallis was used for statistical testing of non-normally distributed data. Red pluses indicate medians.
For the validation study, we used PBMC collected as part of a parent case-control study of TB recurrence in people living with HIV (Supplementary Figure 2, Supplementary Methods). Each episode of TB was bacteriologically confirmed by positive sputum TB GeneXpert, positive sputum smear for acid-fast bacilli, or sputum culture that grew M. tuberculosis. Each participant had completed treatment for TB at least 6 months prior to study enrollment. Recurrent TB was defined as at least 2 episodes of bacteriologically confirmed TB, with the second episode occurring at least 6 months after completing treatment for the first episode. Latent TB testing was performed using the Quantiferon-TB Gold Plus In-Tube interferon-gamma release assay (IGRA) (Qiagen). Blood was collected using EDTA as an anticoagulant for the isolation of plasma and PBMCs. The nested case-control study included all study participants with adequate PBMC available in New York as of October 2021. We calculated that 13 people in the TB group and 13 people in the no-TB group would give 90% power to detect the difference in proviral loads between the TB and no-TB groups seen in the identification subcohort, with a standard deviation of 500. Study participants were enrolled and PBMCs were collected between December 2019 and October 2020.
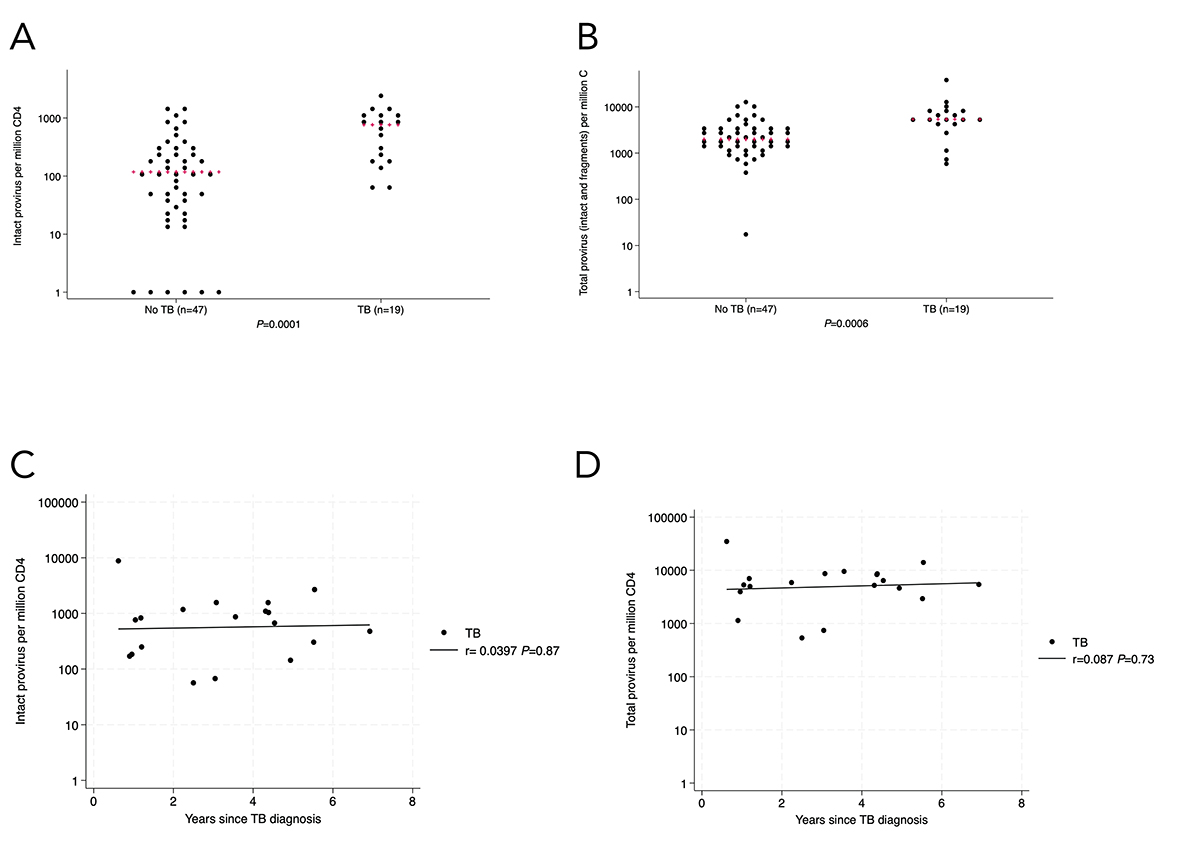
Figure 2. In the analyzed identification cohort subset (n=66), there was a larger number of circulating CD4+ T cells with intact HIV provirus (2A) and total provirus (2B) in people living with HIV with TB or TB history. There was no statistically significant association between intact (2C) or total (2D) provirus in circulating CD4+ T cells and length of time between the last episode of TB and enrollment in the study. Kruskal-Wallis was used for statistical testing of non-normally distributed data. Red pluses indicate medians. Correlations are Pearson’s correlation coefficients.
For both studies, PBMCs were isolated at GHESKIO using density gradient separation (Ficoll-Hypaque, GE Healthcare) with 5x106 PBMC aliquots frozen and then shipped to New York where they were stored at -135C. PBMC specimens were identified with a unique patient identifier that did not correspond to their TB status. Participants were assigned to TB and no-TB groups after laboratory assessments were complete.
For each participant, a single aliquot of 5x106 PBMC was thawed and washed before CD4+ T cell isolation by negative selection (EasySepTM Human CD4+ T cell Enrichment Kit, Stem Cell Technologies). DNA was then isolated using PureGene (Qiagen) for the identification cohort and DNeasy Miniprep (Qiagen) for the validation cohort. Intact proviral DNA amplification was determined using droplet digital PCR (ddPCR) on a QX200 instrument (Bio-Rad) [19]. Amplification of the HIV psi and env regions was performed independently in parallel with quadruple technical replicates. The degree of DNA shearing was assessed using rpp30. Samples with PCR reactions with fewer than 10,000 droplets read were excluded from the calculation for that participant. Samples that did not have any detectable env or psi amplification using the primers and conditions described in Bruner et al [19] and Gunst et al [31] were re-run with secondary env primers and probes as described in Kinloch et al [21]. The rates of primer usage by group are included in Tables 1, 2, and 3. Samples with no amplifiable HIV DNA (env fragment and psi fragment) with both primer sets were also excluded. From the ddPCR, we generated numbers of 3ʹ defective (env), 5ʹ defective (psi), and presumed intact provirus containing both env and psi per million CD4+ T cells.
None of the participants in the identification cohort had detectable HIV in their plasma, but since blood samples from participants in the identification TBRU study were collected with heparin as the anticoagulant, viral load results were potentially not reliable [32]. Therefore, we also tested plasma for circulating HIV p24 GAG protein using an enzyme-linked immunosorbent assay (RETRO-TEK™ HIV-1 p24 Antigen ELISA, ZeptoMetrix) with immune dissociation and reactive confirmation (HIV-1 p24 ICx/CRx kit, ZeptoMetrix). Participants who had detectable p24 or who did not have plasma available for p24 testing were excluded from final analysis of the identification cohort.
Participants in the TB Recurrence study from which the validation cohort was selected were to have had undetectable viral loads prior to enrollment in the study. However, because of the impact that active HIV replication could have on the IPDA, we measured HIV load as well as p24 in these participants. Plasma was centrifuged at 10,000g for 10 minutes to remove particulates. RNA was extracted from plasma using a semi-automated method, and HIV viral load was quantitated using a previously described integrase single copy assay [33, 34]. Samples were analyzed on an ABI Viia7 Real-Time PCR System (Thermo Fisher Scientific). Cycle threshold values were compared with a validated HIV RNA standard run to determine concentration. Participants who had a viral load greater than 1,000 copies/mL and detectable p24 after immune dissociation and neutralization were excluded.
Plasma levels of granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon gamma (IFN-γ), interleukin-1 beta (IL-1β), interleukin-2 (IL-2), interleukin-4 (IL-4), interleukin-5 (IL-5), interleukin-6 (IL-6), interleukin-12p70 (IL-12p70), interleukin-13 (IL-13), interleukin-18 (IL-18), and tumor necrosis factor (TNF) were measured using a bead-based multiplex assay (Th1/Th2 11-plex Human ProcartaPlex Panel, Invitrogen). Quantitation was performed using a Luminex MAGPIX system with xPONENT v4.3 software (Luminex) according to the manufacturer’s instructions with the following specifications. Plasma was centrifuged at 10,000g for 10 minutes to remove particulates prior to aliquoting onto the Luminex plate. Samples were incubated overnight at room temperature. All samples were run in duplicate, and the average reading was used for quantitation with the standard curve. For samples that were less than the lower limit of detection, we imputed a value of half the difference between zero and the lowest concentration standard on the standard curve for each cytokine. Cytokines that were less than the level of detection for more than 50% of plasma samples did not continue to statistical analyses.
Demographic and clinical characteristics were expressed as numbers and percentages with interquartile range given for continuous variables. Time since diagnosis of HIV, time between diagnosis and ART, and time since diagnosis of last episode of TB were converted from days to years for analysis. All statistical analysis was done in Stata (version 18). Distributions of variables were tested using Skewness/Kurtosis tests for normality. For normality test P< 0.05, Kruskal-Wallis tests were used to compare medians. Medians are reported with interquartile range (25th percentile – 75th percentile) as the indicator of variation in non-normally distributed data. Two-sided t tests were used for comparison of means when the normality test P>0.05. For log10 analyses, we added 1 to the proviral load so that the proviral loads of zero would not be undefined (log100 is undefined whereas log101 is zero). When possible, we used the unmodified provirus quantification for statistical calculations. When there was a large number of undetectable proviral loads, we utilized tobit regression for left censoring of proviral loads that were less than the lower limit of detection with the IPDA [35]. For the validation cohort analysis, we completed 2 multivariate regression analyses for factors known to be associated with proviral load and which approached a statistically significant difference between the cases and controls. We used Pearson’s correlation coefficient (<pwcorr> in Stata) to compare quantities of provirus and cytokines. For the cytokine analyses, we applied a Bonferroni correction for multiple comparisons using the number of analyzable cytokines as the correction factor. For all other analyses, the cutoff for significance was P≤0.05.
Using IPDA, we measured intact provirus, psi fragment, and env fragment levels in CD4+ T cells of 100 people living with HIV who had participated in a TB study at GHESKIO Centers in Port au Prince, Haiti. The nested cohort consisted of 25 people with active TB, 25 people with a history of active TB, 25 people with no history of TB and positive IGRA, and 25 people with no history of TB and a negative IGRA. (Table 1, Supplementary Figure 1). This resulted in 50 people in the TB group (25 active + 25 past) and 50 people in the no-TB group (25 IGRA positive + 25 IGRA negative). Despite incidence density matching, the groups had statistically sigificant differences in several factors because of the demographics of the original TBRU cohort (older no-TB controls with a longer time with HIV).
Table 1. Demographics of the Identification Cohort
|
TB (n=50) |
No TB (n=50) |
||||
|
Untreated TB (n=25) |
History of TB or on treatment (n=25) |
IGRA- |
IGRA- |
P- |
|
|
Sex (#female [%]) |
10 (40%) |
14 (56%) |
16 (64%) |
17 (68%) |
0.07 |
|
Age in years (mean +/- SD) |
38.16 +/- 7.5 |
38.8 +/- 6.8 |
44 +/- 5.9 |
44 +/- 4.3 |
0.0001 |
|
Time since HIV dx in years |
0.02 |
3.1 |
12.8 |
11 |
0.0001 |
|
Time on ART in years (median [IQR]) |
0 |
2.9 |
10 |
8.9 |
0.0001 |
|
Time between diagnosis and ART (median [IQR]) |
.06 |
0.9 |
2.7 |
2.6 |
0.0001 |
|
Alternate IPDA primers used (number [%]) |
7 |
9 |
4 |
3 |
0.033 |
|
Time since last diagnosis of TB in years (median [IQR]) |
0.01 |
2.37 |
|||
|
Time between diagnosis of HIV and last diagnosis of TB in years (median [IQR]) |
0.011 |
0.049 |
|||
*P-value is the comparison between TB group (n=50) and no-TB group (n=50) using chi-square (sex), two-sided t test (age), or Mann-Whitney test for non-normally distributed data.
Within the total cohort, there was a greater percentage of CD4+ T cells containing intact provirus in people with current TB or a history of TB vs no TB (median 881 (IQR, 205-2060) vs 116 (IQR, 24-279) copies per million CD4+ T cells, respectively; P=0.0001) (Figure 1A). Tobit regression of detectable intact provirus also showed a statistically significant difference in intact provirus (P < 0.0001) between the TB and no-TB groups. The quantity of psi defective, env defective, total defective, and any viral fragment levels in CD4+ T cells was also statistically significantly higher in the TB group compared to the no-TB group (Figure 1B, Supplementary Figure 3A-C). There was no statistically significant difference in intact provirus between people with newly diagnosed untreated TB (n=25) vs history of TB or TB currently being treated (n=25) (median 897; IQR, 205-3351 vs 865; IQR, 248-1560) copies per million CD4+ T cells, respectively; P=0.92; Figure 1C). In the people with no history of TB, the amount of intact provirus in CD4+ T cells was not statistically significantly different by IGRA status (median 80; IQR, 35-255 for IGRA-positive vs 118; IQR, 21-279 for IGRA-negative per million CD4+ T cells,, P=0.65; Figure 1D). Since there was a statistically significant higher rate of alternate primer usage for IPDA in the TB vs no-TB group (32% vs 14%, respectively), we completed a post-hoc analysis where we stratified by primer type and compared intact proviral load between TB and no-TB groups. The difference in proviral load by TB history status was statistically significant in samples that were amplified with the original primers [19] (P=0.0012) and alternate primers [21] (P=0.016).
To limit potential confounders on the difference in intact provirus observed in the initial groups, we excluded 27 people who had been diagnosed with HIV less than one year before PBMC collection. We excluded 3 people with no plasma available for p24 testing and 3 people with detectable plasma p24. One person who had TB prior to HIV diagnosis was also excluded. All other participants had TB coincident with or after diagnosis of HIV. After these adjustments (Table 2), the difference in intact proviral load between TB and no-TB groups remained statistically significant (median 762; IQR, 183-1173 vs 117; IQR, 24-279 intact provirus per million CD4+ T cells, respectively; P=0.0001) by Kruskal-Wallis test as well as by tobit regression (P<0.0001, Figure 2A). Proviral psi and env fragments and total proviral DNA were also statistically significantly higher in the TB group (Figure 2B, Supplementary Figure 3D-F). There was no association between intact (r= -0.0397, P=0.87, Figure 2C) or total (r=0.087, P=0.73, Figure 2D) provirus in CD4+ T cells and the time since TB diagnosis in the TB group.
For the validation study, we began with 34 participants from a study of TB recurrence. One participant’s PBMC did not have enough live cells after thawing to have CD4+ T cells selected and DNA extracted and was therefore excluded from downstream analyses. One DNA sample failed quality control with no detectable intact env fragment or psi fragment with all tested primer sets and was therefore excluded. One person had HIV viral load >1,000 copies/mL and detectable p24 after dissociation. This person was excluded as the high-level viremia could have rendered the IPDA quantitation inaccurate.
Table 2. Demographics of the Final Analyzed Group in the Identification Cohort
|
TB (n=19) |
No TB (n=47) |
P-value |
|
|
Sex (#female [%]) |
11 (58%) |
30 (63%) |
0.66 |
|
Age (mean [SD]) |
40 (33-47) |
44 (39-49) |
0.005 |
|
Time since HIV dx in years (median [IQR]) |
4.3 (2.3-5.8) |
12.5 (10.6-14.6) |
0.0001 |
|
Time on ART in years (median [IQR]) |
3 (2-5) |
9.3 (8.2-10.4) |
0.0001 |
|
Time between diagnosis and ART (median [IQR]) |
0.13 (0.04-0.9) |
2.6 (1.6-4.2) |
0.0001 |
|
Alternate IPDA primers used (number [%]) |
9 (47%) |
7 (15%) |
0.006 |
|
Time since last diagnosis of TB in years (median [IQR]) |
3.2 (1.18-4.53) |
||
|
Time between diagnosis of HIV and last diagnosis of TB in years (median [IQR]) |
0.112 (0.01-1.76) |
In the final analysis group from the validation cohort (Supplementary Figure S2, Table 3), the quantity of intact provirus was higher in the TB group compared with the no-TB group (median 102; IQR, 0-737 vs 0; IQR, 0-24.5) intact provirus per million CD4+ T cells, respectively P=0.03, Figure 3A). The quantity of total provirus (intact + fragment) was also higher in the TB group compared with the no-TB group (P=0.013, Figure 3B), although the differences in fragment provirus were not statistically significant between the groups (Supplementary Figure 3G-I). We also analyzed the intact provirus levels using a tobit regression model with 12 left-censored observations for intact proviral loads less than the lower limit of detection and the difference remained statistically significant (P=0.04).
Table 3. Demographics of the Final Analyzed Group in the Validation Cohort
|
TB (n=18) |
No TB (n=13) |
P-value |
|
|
Sex (#female [%]) |
10 (55%) |
37 (54%) |
0.9250 |
|
Age (mean [SD]) |
44 +/- 10.1 |
43.3 +/- 12.1 |
0.8626 |
|
Time since HIV dx in years (median [IQR]) |
6.7 (5.4, 7.8) |
6.4 (4.7, 7.9) |
0.8101 |
|
Time on ART in years (median [IQR]) |
6.2 (4.1, 7) |
5 (4, 6.5) |
0.8727 |
|
Time between diagnosis and ART (median [IQR]) |
0.1 (0, 0.2) |
0.8 (0, 1.6) |
0.0722 |
|
HIV viral load (median, [IQR]) |
0 (0-518) |
0 (0-77) |
0.2941 |
|
IGRA positivity (# positive [%]) |
9 (50%) |
10 (77%) |
0.129 |
|
Alternate IPDA primers used (number [%]) |
4 (22.2%) |
3 (23%) |
0.9560 |
|
Time since last diagnosis of TB in years (median [IQR]) |
3.2 (2.3, 5.0) |
||
|
Time between diagnosis of HIV and last diagnosis of TB in years (median [IQR]) |
2.3 (0.03, 4.96) |
||
|
Number with recurrent TB (%) |
7 (38%) |
We conducted multivariable analyses to account for the potential confounders affecting the relationship between a history of TB and intact proviral load. When sex was included in the model as an independent variable [36, 37], the association between TB and proviral load remained statistically significant (P=0.033 for intact provirus, P=0.015 for total provirus). When time since HIV diagnosis and initiation of ART and time between HIV diagnosis and the initiation of ART were included in a model of the association between history of TB and intact proviral load, the association remained statistically significant (P=0.019 and P=0.035, respectively). This was also true with total (intact and defective) provirus as the dependent variable (P=0.018 and P=0.039, respectively). Because the correlation between time with HIV and interval between HIV and ART was significant, we did not generate a multivariate model that included both factors. As the time since HIV diagnosis increased, so did the time between diagnosis and initiation of ART (r = 0.476, P=0.007, Figure 3C), reflecting the evolving recommendations regarding the ideal timeframe to initiate ART over the past 20 years (Figure 3D) [38, 39]. There was no statistically significant correlation between quantity of intact provirus and time interval between HIV diagnosis and ART initiation (r=-0.097, P=0.433; r=0.016, P=0.896, Figure 3E).
There was no statistically significant correlation between intact provirus and plasma cytokine levels for the validation cohort. However, we found that the frequency of CD4+ T cells with any detectable provirus (intact + fragment) was directly proportional to the levels of IL-1β (r = 0.524, P=0.0025), IL-2 (r = 0.622, P=0.0002), IL-12p70 (r = 0.491, P=0.0093), and IL-13 (r = 0.4769, P=0.0067). After Bonferroni correction for the 8 analyzable cytokines, (cutoff Padj=0.05/8=0.006), the association between plasma IL-1β (Figure 4A) and plasma IL-2 (Figure 4B) with CD4+ T cells containing HIV env or psi remained statistically significant.
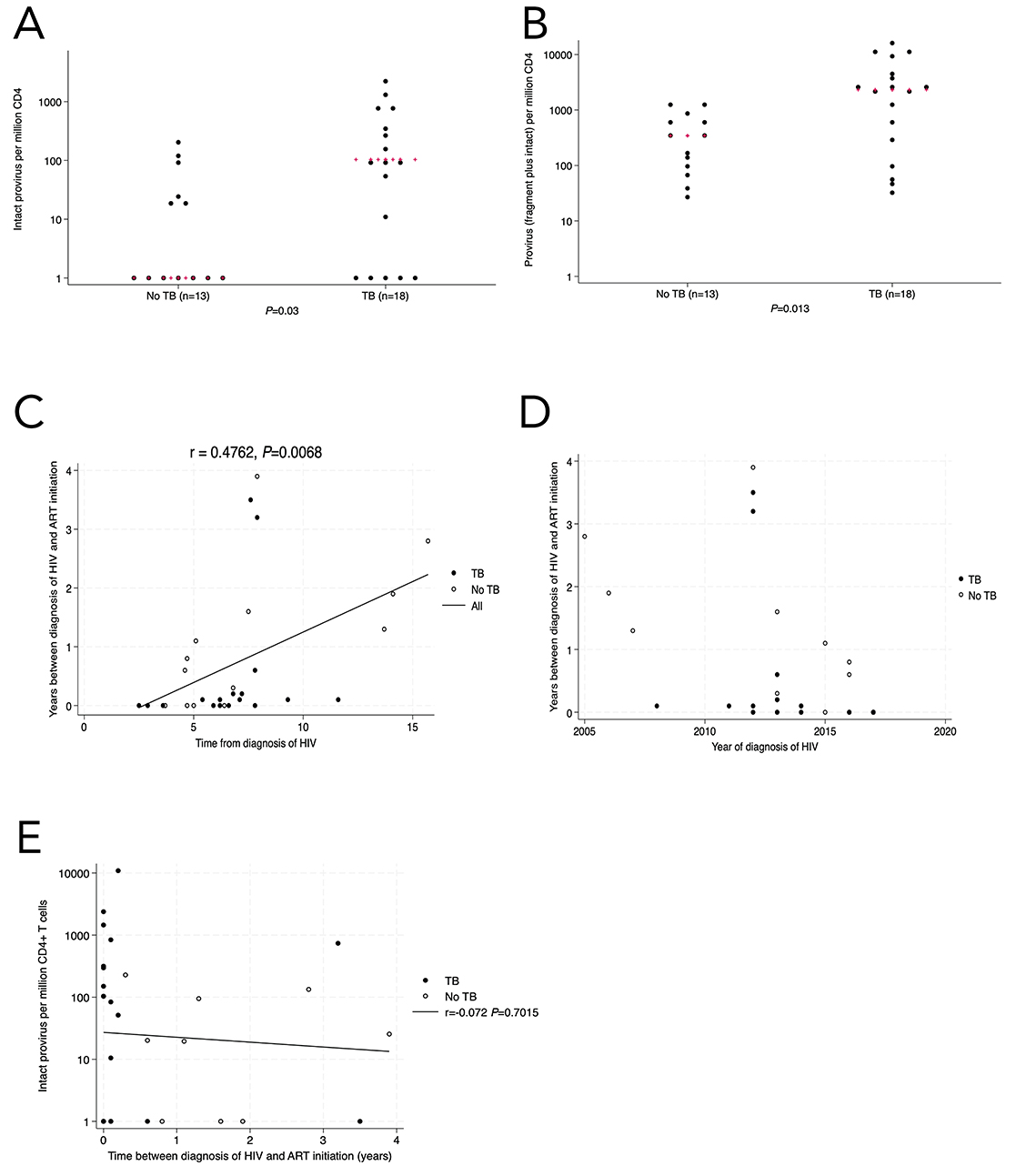
Figure 3. In the validation cohort (n=31), there was a higher level of intact (3A) and total provirus (3B) in the TB group compared with people with no history of active pulmonary TB. When completing multivariate analysis, we found a statistically significant correlation between time since HIV diagnosis (x-axis) and the duration of time between HIV diagnosis and ART initiation (y-axis) (3C), which was likely related to changing recommendations of when to start ART relative to CD4+ T cell count (3D). There was no statistically significant correlation between time without ART and proviral load (3E). Kruskal-Wallis was used for statistical testing of non-normally distributed data. Red pluses are the medians. Correlations are Pearson’s correlation coefficients.
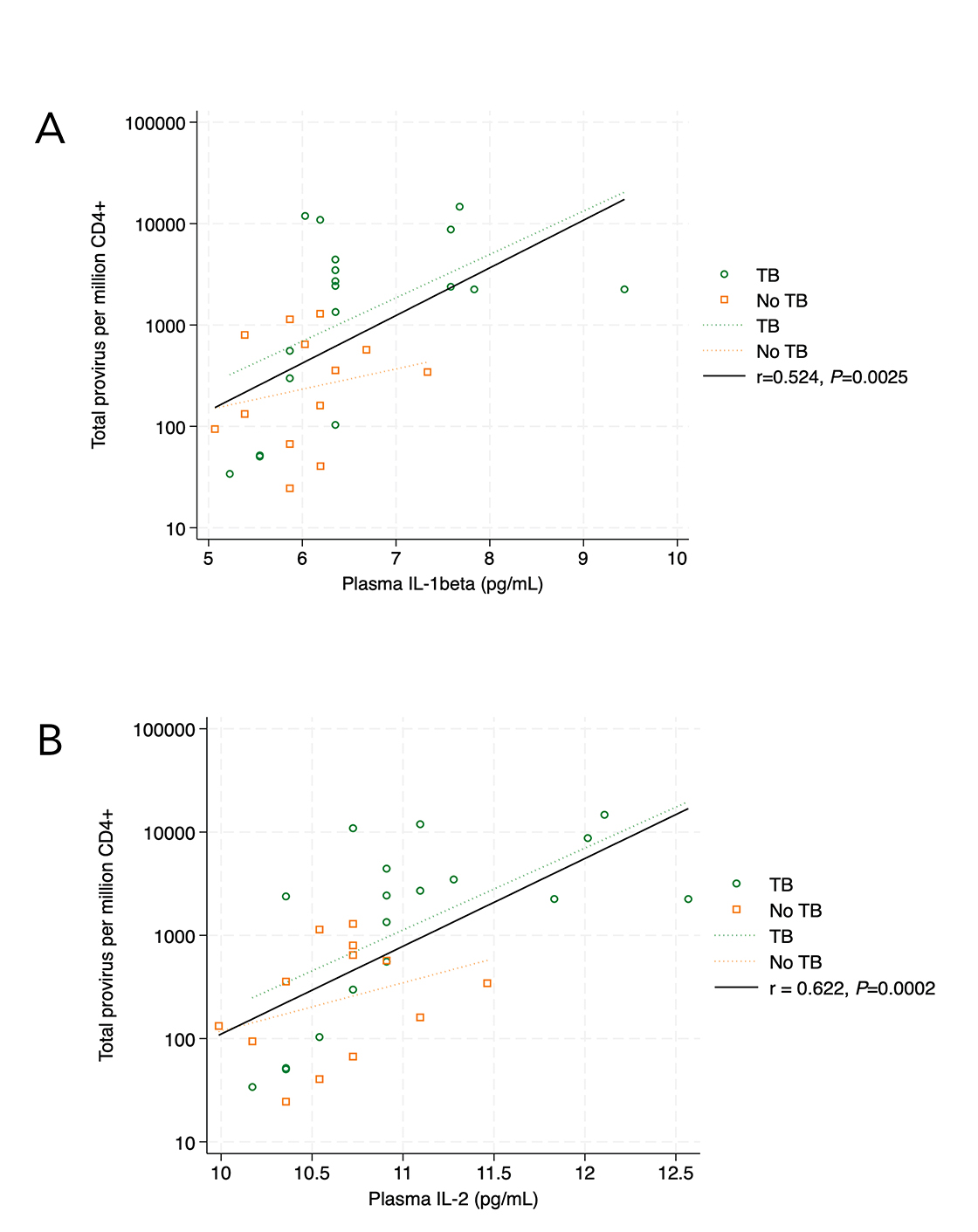
Figure 4. In the validation cohort, plasma IL-1β levels (4A) correlated with circulating CD4+ T cells containing HIV psi and/or env (intact + fragment) (r = 0.524, P=0.0025, 4A). The same association was found for plasma IL-2 levels and circulating CD4+ T cells containing HIV psi and/or env (intact + fragment) (r = 0.622, P=0.0002, 4B). Correlations are Pearson’s correlation coefficients.
Using IPDA, we documented significantly higher levels of intact, fragment, and total HIV proviruses in circulating CD4+ T cells of people living with HIV and a history of bacteriologically confirmed pulmonary TB. The larger percentage of CD4+ T cells containing intact HIV provirus in people with a history of TB was shown in an identification cohort and confirmed in a validation cohort. The statistically significant difference persisted when controlling for important potential confounders of increased proviral load including time since HIV diagnosis, interval of time between HIV diagnosis and ART initiation, and sex. Interestingly the percentage of CD4+ T cells containing intact provirus was not correlated with time since last TB diagnosis; this suggests that differential reservoir size persists even after TB cure, although longitudinal studies are needed to assess this hypothesis.
T cells differentiate and expand differently in latent and active TB [40], with much higher levels of activated CD4+ T cells during active TB. Studies using PBMC from people with latent TB without HIV have found CD4+ T cells with common T cell receptor-beta sequences for different M. tuberculosis antigenic epitopes in people who do vs do not progress to active TB [41]. Characterization of CD4+ T cell phenotype and antigen specificity in people with HIV at various stages of HIV found more clonality of T cells and infecting provirus during later stages of HIV. In a study of T cell receptors of HIV-infected CD4+ T cells from 17 people, they found that 2 people had common anti-M. tuberculosis epitopes on CD4+ T cells, but that M. tuberculosis-specific CD4+ T cells were not over-represented in the overall p24+ CD4+ T cell pool [42]. While our results indicate a higher intact and total proviral load in people with a history of pulmonary TB disease, future studies will include characterization of which types of CD4+ T cells and which antigen specificities contain provirus in their DNA [43]. A 2010 study concluded that M. tuberculosis-specific CD4+ T cells were more likely to be infected by HIV, but were also more likely to be depleted during the subsequent course of infection [44]. The responsiveness to M. tuberculosis was based on cell surface marker phenotypes and cytokine production in reponse to M. tuberculosis “antigens.” Newer methodologies will allow us to determine CD4+ T cell reactivity at the level of the T cell receptor epitope in cells infected with HIV.
Local inflammation can increase HIV replication and viral load at sites where HIV and replicating M. tuberculosis exist, even prior to the development of symptomatic TB [45]. The increase in inflammation in people with latent TB that precedes clinically apparent active TB disease [46] may drive the increase in the HIV reservoir by activating latently infected cells or generating more activated cells that are at risk for infection by circulating HIV. The existing literature is inconclusive about a correlation between HIV provirus and inflammation [37, 47, 48] and TB and inflammation [49–51]. This study’s contribution is in demonstrating a correlation of total provirus with larger plasma concentrations of pro-inflammatory cytokines IL-1β and IL-2. Production of these cytokines may drive HIV activation or be a response to HIV [52]; they are also important in the balance between control of M. tuberculosis infection and pathology related to immune response to M. tuberculosis. [53]
A strength of this study is the assessment of intact provirus in CD4+ T cells of people living with HIV in a resource-limited setting where TB is syndemic. The use of IPDA to quantitate provirus from a relatively small volume of starting material (5x106 PBMC) meant that PBMC collected from standard phlebotomy during a research cohort study could be leveraged. This has important implications for how people can use IPDA to study HIV reservoir in a variety of research and clinical settings. Study limitations include incomplete nadir CD4+ T cell count and HIV viral load testing due to the changing landscape of CD4+ T cell and viral load testing in HIV care and the variability in the availability of this test at GHESKIO. This study only included people with bacteriologically confirmed pulmonary TB in order to have a TB group with highest certainty for having had TB. However, we do not know how these results will be generalizable to people treated empirically for TB or with extrapulmonary TB. We hypothesize that we will see the same increase in circulating CD4+ T cell HIV reservoir, but that study needs to be conducted. We note that the prevalent HIV subtype in Haiti is subtype B, and that this study needs to be replicated in communities where subtypes other than subtype B are prevalent. In addition to differences in techniques for quantitating HIV provirus, different HIV subtypes might explain the discrepant results regarding association between HIV DNA levels and TB seen in studies from China, Uganda, and now Haiti [14, 15].
We used 2 sets of primers to amplify HIV provirus from CD4+ T cells, which is consistent with other studies using clinical samples from non-US locations [21]. The percentage of DNA in the TB and no-TB groups needing alternate primers for amplification was statistically significantly different in the identification cohort, which may reflect different times of HIV acquisition in the TB and no-TB cohorts. When comparing proviral load obtained using original or alternate primers, the association between TB and HIV proviral load remained statistically significant. The median intact proviral loads were approximately 7-fold higher in the TB group compared with the no-TB group in both the identification and validation cohorts. However, the median intact proviral loads for the groups differed between the identification and validation cohorts. This may be because of differences in the parent studies for the identification and validation cohorts or that the testing of the identification and validation cohorts was separated by over a year which meant different reagent lot numbers, different technicians running the assay, and a different standardized DNA extraction method in use in the lab. We believe this underscores the importance of running appropriate controls for every set of IPDA experiments.
In the identification cohort, we did not see a statistically significant difference in intact provirus between IGRA-positive and IGRA-negative people living with HIV who did not have a history of active TB. Everyone diagnosed with HIV at GHESKIO is treated empirically for latent TB, so all of these participants would have received isoniazid since diagnosis of HIV [27]. IGRA reversion rates after latent TB treatment are not indicative of treatment success or failure [54]. Participants in these 2 cohorts live in a region where the rate of new diagnosis of pulmonary TB was 5% in a recent community-level screening [55] with a rate of 70% IGRA positivity in people without HIV [28]. Therefore, regardless of the result of the IGRA, we know that study participants in care at GHESKIO have a high baseline level of exposure to M. tuberculosis. Participants in the validation cohort TB and no-TB groups were well-matched with regard to age, sex, duration of HIV, and time between HIV diagnosis and ART initiation, with one group mounting an ex vivo interferon gamma response to M. tuberculosis-specific peptides as measured by IGRA and the other not. Therefore, we suggest that the development of and survival after active TB disease may be the driver of increased provirus load in circulating CD4+ T cells, rather than M. tuberculosis infection, per se. Of note, the participants in the identification cohort were tested for latent TB using the Quantiferon Gold assay, which did not yet include a peptide panel optimized for recognition by CD8+ T cells.
The increased risk of TB for people living with HIV does not normalize to that of the general population even when HIV is virologically suppressed and CD4+ T cell counts have recovered. People with TB have higher all-cause mortality even after TB is cured, and people with higher HIV reservoir have worse clinical outcomes. Therefore, we hypothesized that HIV reservoir size would be higher in people with a history of TB, and this is supported by our data showing increased intact and total provirus in circulating CD4+ T cells of people with a history of bacteriologically confirmed active pulmonary TB in 2 different cohorts in Haiti. TB and other coinfections should be considered when studying the dynamics of the HIV reservoir, particularly when evaluating candidate HIV cure strategies.
We acknowledge the patient care, clinical research, and laboratory teams at GHESKIO Centers in Port au Prince, Haiti and all of our study participants. We especially thank the GHESKIO laboratory team members who processed blood for peripheral blood mononuclear cell isolation and storage, without whom this study would not have been possible.
The authors recognize funding to KD (NIH K23-AI131913 and R01-AI176943, Doris Duke Charitable Foundation Clinical Scientist Development Award) and DWF (NIH U19-AI111143, P30-AI168433, and K24-AI098627). Additionally, this project utilized databases maintained in REDCap, supported by UL1-TR002385 from the National Center for Advancing Translational Sciences of the NIH.
The authors report no competing financial interests.
Supplementary materials are available at the Pathogens and Immunity website. Supplementary data may be provided by the authors to benefit the reader. Supplementary data are not copyedited and are the sole responsibility of the authors. Questions or comments related to supplementary materials should be addressed to the corresponding author.
1. de Oliveira MF, Murrell B, Perez-Santiago J, Vargas M, Ellis RJ, Letendre S, Grant I, Smith DM, Woods SP, Gianella S. Circulating HIV DNA Correlates With Neurocognitive Impairment in Older HIV-infected Adults on Suppressive ART. Sci Rep. 2015;5:17094. doi: 10.1038/srep17094. PubMed PMID: 26603568; PMCID: PMC4658529.
2. Ruhanya V, Jacobs GB, Glashoff RH, Engelbrecht S. Clinical Relevance of Total HIV DNA in Peripheral Blood Mononuclear Cell Compartments as a Biomarker of HIV-Associated Neurocognitive Disorders (HAND). Viruses. 2017;9(11). doi: 10.3390/v9110324. PubMed PMID: 29088095; PMCID: PMC5707531.
3. Rouzioux C, Hubert JB, Burgard M, Deveau C, Goujard C, Bary M, Sereni D, Viard JP, Delfraissy JF, Meyer L, Group SCS. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192(1):46-55. doi: 10.1086/430610. PubMed PMID: 15942893.
4. Goujard C, Bonarek M, Meyer L, Bonnet F, Chaix ML, Deveau C, Sinet M, Galimand J, Delfraissy JF, Venet A, Rouzioux C, Morlat P, Agence Nationale de Recherche sur le Sida PSG. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis. 2006;42(5):709-15. doi: 10.1086/500213. PubMed PMID: 16447119.
5. Abrahams MR, Joseph SB, Garrett N, Tyers L, Moeser M, Archin N, Council OD, Matten D, Zhou S, Doolabh D, Anthony C, Goonetilleke N, Karim SA, Margolis DM, Pond SK, Williamson C, Swanstrom R. The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci Transl Med. 2019;11(513). doi: 10.1126/scitranslmed.aaw5589. PubMed PMID: 31597754; PMCID: PMC7233356.
6. Boulassel MR, Chomont N, Pai NP, Gilmore N, Sekaly RP, Routy JP. CD4 T cell nadir independently predicts the magnitude of the HIV reservoir after prolonged suppressive antiretroviral therapy. J Clin Virol. 2012;53(1):29-32. doi: 10.1016/j.jcv.2011.09.018. PubMed PMID: 22019250.
7. Massanella M, Bender Ignacio RA, Lama JR, Pagliuzza A, Dasgupta S, Alfaro R, Rios J, Ganoza C, Pinto-Santini D, Gilada T, Duerr A, Chomont N, Group MS. Long-term effects of early antiretroviral initiation on HIV reservoir markers: a longitudinal analysis of the MERLIN clinical study. Lancet Microbe. 2021;2(5):e198-e209. doi: 10.1016/S2666-5247(21)00010-0. PubMed PMID: 35544209.
8. Reddy K, Lee GQ, Reddy N, Chikowore TJB, Baisley K, Dong KL, Walker BD, Yu XG, Lichterfeld M, Ndung’u T. Differences in HIV-1 reservoir size, landscape characteristics and decay dynamics in acute and chronic treated HIV-1 Clade C infection. medRxiv. 2024. doi: 10.1101/2024.02.16.24302713. PubMed PMID: 38947072; PMCID: PMC11213047.
9. Stevenson EM, Terry S, Copertino D, Leyre L, Danesh A, Weiler J, Ward AR, Khadka P, McNeil E, Bernard K, Miller IG, Ellsworth GB, Johnston CD, Finkelsztein EJ, Zumbo P, Betel D, Dundar F, Duncan MC, Lapointe HR, Speckmaier S, Moran-Garcia N, Papa MP, Nicholes S, Stover CJ, Lynch RM, Caskey M, Gaebler C, Chun TW, Bosque A, Wilkin TJ, Lee GQ, Brumme ZL, Jones RB. SARS CoV-2 mRNA vaccination exposes latent HIV to Nef-specific CD8(+) T-cells. Nat Commun. 2022;13(1):4888. doi: 10.1038/s41467-022-32376-z. PubMed PMID: 35985993; PMCID: PMC9389512.
10. Jones RB, Kovacs C, Chun TW, Ostrowski MA. Short communication: HIV type 1 accumulates in influenza-specific T cells in subjects receiving seasonal vaccination in the context of effective antiretroviral therapy. AIDS Res Hum Retroviruses. 2012;28(12):1687-92. doi: 10.1089/AID.2012.0115. PubMed PMID: 22734882; PMCID: PMC3505056.
11. Gianella S, Anderson CM, Var SR, Oliveira MF, Lada SM, Vargas MV, Massanella M, Little SJ, Richman DD, Strain MC, Perez-Santiago J, Smith DM. Replication of Human Herpesviruses Is Associated with Higher HIV DNA Levels during Antiretroviral Therapy Started at Early Phases of HIV Infection. J Virol. 2016;90(8):3944-52. doi: 10.1128/JVI.02638-15. PubMed PMID: 26842469; PMCID: PMC4810527.
12. Gianella S, Tran SM, Morris S, Vargas M, Porrachia M, Oliveira MF, Lada S, Zhao M, Ellsworth GB, Mathad JS, Wilkin T. Sex Differences in CMV Replication and HIV Persistence During Suppressive ART. Open Forum Infect Dis. 2020;7(8):ofaa289. doi: 10.1093/ofid/ofaa289. PubMed PMID: 32793766; PMCID: PMC7415302.
13. Organization WH. WHO consolidated guidelines on tuberculosis 2024 [cited Module 6: Tuberculosis and comorbidities June 28, 2024]. Available from: https://iris.who.int/bitstream/handle/10665/376584/9789240087002-eng.pdf?sequence=1.
14. Olson A, Ragan EJ, Nakiyingi L, Lin N, Jacobson KR, Ellner JJ, Manabe YC, Sagar M. Brief Report: Pulmonary Tuberculosis Is Associated With Persistent Systemic Inflammation and Decreased HIV-1 Reservoir Markers in Coinfected Ugandans. J Acquir Immune Defic Syndr. 2018;79(3):407-11. doi: 10.1097/QAI.0000000000001823. PubMed PMID: 30063648; PMCID: PMC6506856.
15. Xun J, Qi T, Zou L, Tang Q, Shen Y, Yang J, Xie L, Ji Y, Zhang R, Liu L, Wang J, Steinhart C, Wang Z, Tang Y, Song W, Sun J, Cheng J, Le X, Wu H, He X, Chen R, Chen J, Lu H. Mycobacterium tuberculosis co-infection is associated with increased surrogate marker of the HIV reservoir. AIDS Res Ther. 2020;17(1):63. doi: 10.1186/s12981-020-00320-0. PubMed PMID: 33076959; PMCID: PMC7574250.
16. Goletti D, Weissman D, Jackson RW, Graham NM, Vlahov D, Klein RS, Munsiff SS, Ortona L, Cauda R, Fauci AS. Effect of Mycobacterium tuberculosis on HIV replication. Role of immune activation. J Immunol. 1996;157(3):1271-8. PubMed PMID: 8757635.
17. Toossi Z, Mayanja-Kizza H, Hirsch CS, Edmonds KL, Spahlinger T, Hom DL, Aung H, Mugyenyi P, Ellner JJ, Whalen CW. Impact of tuberculosis (TB) on HIV-1 activity in dually infected patients. Clin Exp Immunol. 2001;123(2):233-8. doi: 10.1046/j.1365-2249.2001.01401.x. PubMed PMID: 11207653; PMCID: PMC1905977.
18. Falcinelli SD, Ceriani C, Margolis DM, Archin NM. New Frontiers in Measuring and Characterizing the HIV Reservoir. Front Microbiol. 2019;10:2878. doi: 10.3389/fmicb.2019.02878. PubMed PMID: 31921056; PMCID: PMC6930150.
19. Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566(7742):120-5. doi: 10.1038/s41586-019-0898-8. PubMed PMID: 30700913; PMCID: PMC6447073.
20. Simonetti FR, White JA, Tumiotto C, Ritter KD, Cai M, Gandhi RT, Deeks SG, Howell BJ, Montaner LJ, Blankson JN, Martin A, Laird GM, Siliciano RF, Mellors JW, Siliciano JD. Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc Natl Acad Sci U S A. 2020;117(31):18692-700. doi: 10.1073/pnas.2006816117. PubMed PMID: 32690683; PMCID: PMC7414172.
21. Kinloch NN, Ren Y, Conce Alberto WD, Dong W, Khadka P, Huang SH, Mota TM, Wilson A, Shahid A, Kirkby D, Harris M, Kovacs C, Benko E, Ostrowski MA, Del Rio Estrada PM, Wimpelberg A, Cannon C, Hardy WD, MacLaren L, Goldstein H, Brumme CJ, Lee GQ, Lynch RM, Brumme ZL, Jones RB. HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA). Nat Commun. 2021;12(1):165. doi: 10.1038/s41467-020-20442-3. PubMed PMID: 33420062; PMCID: PMC7794580.
22. Joseph Y, Yao Z, Dua A, Severe P, Collins SE, Bang H, Antoine Jean-Juste M, Ocheretina O, Apollon A, McNairy ML, Dupnik K, Cremieux E, Byrne A, Pape JW, Koenig SP. Long-term mortality after tuberculosis treatment among persons living with HIV in Haiti. J Int AIDS Soc. 2021;24(7):e25721. doi: 10.1002/jia2.25721. PubMed PMID: 34235862; PMCID: PMC8264404.
23. Organization WH. Haiti: HIV Country Profile 2023 2023 [June 28, 2024]. Available from: https://cfs.hivci.org/index.html.
24. Organization WH. Tuberculosis profile: Haiti.
25. Daniels JP. GHESKIO: adapting to challenges in Haiti since the early 1980s. Lancet HIV. 2019;6(3):e153. doi: 10.1016/S2352-3018(19)30048-7. PubMed PMID: 30846057.
26. Fitzgerald DW, Marotte C, Verdier RI, Johnson WD, Jr., Pape JW. Comprehension during informed consent in a less-developed country. Lancet. 2002;360(9342):1301-2. doi: 10.1016/S0140-6736(02)11338-9. PubMed PMID: 12414207.
27. Pape JW, Jean SS, Ho JL, Hafner A, Johnson WD, Jr. Effect of isoniazid prophylaxis on incidence of active tuberculosis and progression of HIV infection. Lancet. 1993;342(8866):268-72. doi: 10.1016/0140-6736(93)91817-6. PubMed PMID: 8101302.
28. Dupnik KM, Bean JM, Lee MH, Jean Juste MA, Skrabanek L, Rivera V, Vorkas CK, Pape JW, Fitzgerald DW, Glickman M. Blood transcriptomic markers of Mycobacterium tuberculosis load in sputum. Int J Tuberc Lung Dis. 2018;22(8):950-8. doi: 10.5588/ijtld.17.0855. PubMed PMID: 29991407; PMCID: PMC6343854.
29. Vorkas CK, Wipperman MF, Li K, Bean J, Bhattarai SK, Adamow M, Wong P, Aube J, Juste MAJ, Bucci V, Fitzgerald DW, Glickman MS. Mucosal-associated invariant and gammadelta T cell subsets respond to initial Mycobacterium tuberculosis infection. JCI Insight. 2018;3(19). doi: 10.1172/jci.insight.121899. PubMed PMID: 30282828; PMCID: PMC6237486.
30. Wipperman MF, Fitzgerald DW, Juste MAJ, Taur Y, Namasivayam S, Sher A, Bean JM, Bucci V, Glickman MS. Antibiotic treatment for Tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci Rep. 2017;7(1):10767. doi: 10.1038/s41598-017-10346-6. PubMed PMID: 28883399; PMCID: PMC5589918.
31. Gunst JD, Pahus MH, Rosas-Umbert M, Lu IN, Benfield T, Nielsen H, Johansen IS, Mohey R, Ostergaard L, Klastrup V, Khan M, Schleimann MH, Olesen R, Stovring H, Denton PW, Kinloch NN, Copertino DC, Ward AR, Alberto WDC, Nielsen SD, Puertas MC, Ramos V, Reeves JD, Petropoulos CJ, Martinez-Picado J, Brumme ZL, Jones RB, Fox J, Tolstrup M, Nussenzweig MC, Caskey M, Fidler S, Sogaard OS. Early intervention with 3BNC117 and romidepsin at antiretroviral treatment initiation in people with HIV-1: a phase 1b/2a, randomized trial. Nat Med. 2022;28(11):2424-35. doi: 10.1038/s41591-022-02023-7. PubMed PMID: 36253609; PMCID: PMC10189540.
32. Holodniy M, Kim S, Katzenstein D, Konrad M, Groves E, Merigan TC. Inhibition of human immunodeficiency virus gene amplification by heparin. J Clin Microbiol. 1991;29(4):676-9. doi: 10.1128/jcm.29.4.676-679.1991. PubMed PMID: 1909709; PMCID: PMC269852.
33. Cillo AR, Vagratian D, Bedison MA, Anderson EM, Kearney MF, Fyne E, Koontz D, Coffin JM, Piatak M, Jr., Mellors JW. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol. 2014;52(11):3944-51. doi: 10.1128/JCM.02060-14. PubMed PMID: 25187636; PMCID: PMC4313209.
34. McCann CD, van Dorp CH, Danesh A, Ward AR, Dilling TR, Mota TM, Zale E, Stevenson EM, Patel S, Brumme CJ, Dong W, Jones DS, Andresen TL, Walker BD, Brumme ZL, Bollard CM, Perelson AS, Irvine DJ, Jones RB. A participant-derived xenograft model of HIV enables long-term evaluation of autologous immunotherapies. J Exp Med. 2021;218(7). doi: 10.1084/jem.20201908. PubMed PMID: 33988715; PMCID: PMC8129803.
35. Tobin J. Estimation of Relationships for Limited Dependent Variables. Econometrica. 1958;26(1):24-36. doi: 10.2307/1907382. PubMed PMID: WOS:A1958CAL6600002.
36. Falcinelli SD, Shook-Sa BE, Dewey MG, Sridhar S, Read J, Kirchherr J, James KS, Allard B, Ghofrani S, Stuelke E, Baker C, Roan NR, Eron JJ, Kuruc JD, Ramirez C, Gay C, Mollan KR, Margolis DM, Adimora AA, Archin NM. Impact of Biological Sex on Immune Activation and Frequency of the Latent HIV Reservoir During Suppressive Antiretroviral Therapy. J Infect Dis. 2020;222(11):1843-52. doi: 10.1093/infdis/jiaa298. PubMed PMID: 32496542; PMCID: PMC7653086.
37. Prodger JL, Capoferri AA, Yu K, Lai J, Reynolds SJ, Kasule J, Kityamuweesi T, Buule P, Serwadda D, Kwon KJ, Schlusser K, Martens C, Scully E, Choi YH, Redd AD, Quinn TC. Reduced HIV-1 latent reservoir outgrowth and distinct immune correlates among women in Rakai, Uganda. JCI Insight. 2020;5(14). doi: 10.1172/jci.insight.139287. PubMed PMID: 32544096; PMCID: PMC7453892.
38. Severe P, Juste MA, Ambroise A, Eliacin L, Marchand C, Apollon S, Edwards A, Bang H, Nicotera J, Godfrey C, Gulick RM, Johnson WD, Jr., Pape JW, Fitzgerald DW. Early versus standard antiretroviral therapy for HIV-infected adults in Haiti. N Engl J Med. 2010;363(3):257-65. doi: 10.1056/NEJMoa0910370. PubMed PMID: 20647201; PMCID: PMC3676927.
39. Koenig SP, Dorvil N, Devieux JG, Hedt-Gauthier BL, Riviere C, Faustin M, Lavoile K, Perodin C, Apollon A, Duverger L, McNairy ML, Hennessey KA, Souroutzidis A, Cremieux PY, Severe P, Pape JW. Same-day HIV testing with initiation of antiretroviral therapy versus standard care for persons living with HIV: A randomized unblinded trial. PLoS Med. 2017;14(7):e1002357. doi: 10.1371/journal.pmed.1002357. PubMed PMID: 28742880; PMCID: PMC5526526.
40. Ahmed A, Vyakarnam A. Emerging patterns of regulatory T cell function in tuberculosis. Clin Exp Immunol. 2020;202(3):273-87. doi: 10.1111/cei.13488. PubMed PMID: 32639588; PMCID: PMC7670141.
41. Musvosvi M, Huang H, Wang C, Xia Q, Rozot V, Krishnan A, Acs P, Cheruku A, Obermoser G, Leslie A, Behar SM, Hanekom WA, Bilek N, Fisher M, Kaufmann SHE, Walzl G, Hatherill M, Davis MM, Scriba TJ, Adolescent Cohort Study t, Consortium GC. T cell receptor repertoires associated with control and disease progression following Mycobacterium tuberculosis infection. Nat Med. 2023;29(1):258-69. doi: 10.1038/s41591-022-02110-9. PubMed PMID: 36604540; PMCID: PMC9873565.
42. Gantner P, Buranapraditkun S, Pagliuzza A, Dufour C, Pardons M, Mitchell JL, Kroon E, Sacdalan C, Tulmethakaan N, Pinyakorn S, Robb ML, Phanuphak N, Ananworanich J, Hsu D, Vasan S, Trautmann L, Fromentin R, Chomont N. HIV rapidly targets a diverse pool of CD4(+) T cells to establish productive and latent infections. Immunity. 2023;56(3):653-68 e5. doi: 10.1016/j.immuni.2023.01.030. PubMed PMID: 36804957; PMCID: PMC10023508.
43. Mendoza P, Jackson JR, Oliveira TY, Gaebler C, Ramos V, Caskey M, Jankovic M, Nussenzweig MC, Cohn LB. Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J Exp Med. 2020;217(7). doi: 10.1084/jem.20200051. PubMed PMID: 32311008; PMCID: PMC7336300.
44. Geldmacher C, Ngwenyama N, Schuetz A, Petrovas C, Reither K, Heeregrave EJ, Casazza JP, Ambrozak DR, Louder M, Ampofo W, Pollakis G, Hill B, Sanga E, Saathoff E, Maboko L, Roederer M, Paxton WA, Hoelscher M, Koup RA. Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J Exp Med. 2010;207(13):2869-81. doi: 10.1084/jem.20100090. PubMed PMID: 21115690; PMCID: PMC3005236.
45. Waters R, Ndengane M, Abrahams MR, Diedrich CR, Wilkinson RJ, Coussens AK. The Mtb-HIV syndemic interaction: why treating M. tuberculosis infection may be crucial for HIV-1 eradication. Future Virol. 2020;15(2):101-25. doi: 10.2217/fvl-2019-0069. PubMed PMID: 32273900; PMCID: PMC7132588.
46. Scriba TJ, Penn-Nicholson A, Shankar S, Hraha T, Thompson EG, Sterling D, Nemes E, Darboe F, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Johnson JL, Boom WH, Hatherill M, Valvo J, De Groote MA, Ochsner UA, Aderem A, Hanekom WA, Zak DE, other members of the ACScst. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 2017;13(11):e1006687. doi: 10.1371/journal.ppat.1006687. PubMed PMID: 29145483; PMCID: PMC5689825.
47. Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, Rinaldo CR, Riddler SA, Hogg E, Godfrey C, Collier AC, Eron JJ, Mellors JW, Team AA. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog. 2017;13(4):e1006285. doi: 10.1371/journal.ppat.1006285. PubMed PMID: 28426825; PMCID: PMC5398724.
48. Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, Capoferri AA, Beg SA, Huang SH, Karandish S, Hao H, Halper-Stromberg E, Yong PC, Kovacs C, Benko E, Siliciano RF, Ho YC. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe. 2017;21(4):494-506 e4. doi: 10.1016/j.chom.2017.03.008. PubMed PMID: 28407485; PMCID: PMC5433942.
49. Du Bruyn E, Fukutani KF, Rockwood N, Schutz C, Meintjes G, Arriaga MB, Cubillos-Angulo JM, Tiburcio R, Sher A, Riou C, Wilkinson KA, Andrade BB, Wilkinson RJ. Inflammatory profile of patients with tuberculosis with or without HIV-1 co-infection: a prospective cohort study and immunological network analysis. Lancet Microbe. 2021;2(8):e375-e85. doi: 10.1016/S2666-5247(21)00037-9. PubMed PMID: 35544195.
50. Kusejko K, Gunthard HF, Olson GS, Zens K, Darling K, Khanna N, Furrer H, Vetter P, Bernasconi E, Vernazza P, Hoffmann M, Kouyos RD, Nemeth J, Swiss HIVCS. Diagnosis of latent tuberculosis infection is associated with reduced HIV viral load and lower risk for opportunistic infections in people living with HIV. PLoS Biol. 2020;18(12):e3000963. doi: 10.1371/journal.pbio.3000963. PubMed PMID: 33284802; PMCID: PMC7721132.
51. Gupta A, Wood R, Kaplan R, Bekker LG, Lawn SD. Prevalent and incident tuberculosis are independent risk factors for mortality among patients accessing antiretroviral therapy in South Africa. PLoS One. 2013;8(2):e55824. doi: 10.1371/journal.pone.0055824. PubMed PMID: 23418463; PMCID: PMC3572168.
52. Riitho V, Connon R, Gwela A, Namusanje J, Nhema R, Siika A, Bwakura-Dangarembizi M, Musiime V, Berkley JA, Szubert AJ, Gibb DM, Walker AS, Klein N, Prendergast AJ. Biomarkers of mortality in adults and adolescents with advanced HIV in sub-Saharan Africa. Nat Commun. 2024;15(1):5492. doi: 10.1038/s41467-024-49317-7. PubMed PMID: 38944653; PMCID: PMC11214617.
53. Silverio D, Goncalves R, Appelberg R, Saraiva M. Advances on the Role and Applications of Interleukin-1 in Tuberculosis. mBio. 2021;12(6):e0313421. doi: 10.1128/mBio.03134-21. PubMed PMID: 34809460; PMCID: PMC8609357.
54. Xin H, Cao X, Zhang H, Liu J, Pan S, Li X, Guan L, Shen F, Liu Z, Wang D, Guan X, Yan J, Li H, Feng B, Zhang M, Yang Q, Jin Q, Gao L, Team L-NS. Dynamic changes of interferon gamma release assay results with latent tuberculosis infection treatment. Clin Microbiol Infect. 2020;26(11):1555 e1- e7. doi: 10.1016/j.cmi.2020.02.009. PubMed PMID: 32062048.
55. Rivera VR, Lu L, Ocheretina O, Jean Juste MA, Julma P, Archange D, Moise CG, Homeus F, Phanor PD, Petion S, Cremieux PY, Fitzgerald DW, Pape JW, Koenig SP. Diagnostic yield of active case finding for tuberculosis at human immunodeficiency virus testing in Haiti. Int J Tuberc Lung Dis. 2019;23(11):1217-22. doi: 10.5588/ijtld.18.0835. PubMed PMID: 31718759; PMCID: PMC7647668.
Submitted June 4, 2024 | Accepted August 22, 2024 | Published September 23, 2024
Copyright © 2024 The Authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License.