Wilfried Wenceslas Bazié1,2,3, Julien Boucher1,2, Julien Vitry1,2, Benjamin Goyer1, Jean Pierre Routy4,5, Cécile Tremblay6,7, Sylvie Trottier1,2, Mohammad-Ali Jenabian8, Patrick Provost1,2, Michel Alary1,9,10 and Caroline Gilbert1,2
1Centre de recherche du CHU de Québec-Université Laval, Québec, QC, Canada
2Département de microbiologie-infectiologie et d’immunologie, Faculté de médecine, Université Laval, Québec, QC, Canada
4Programme de recherche sur les maladies infectieuses, Centre Muraz, Institut National de Santé Publique, Bobo-Dioulasso, Burkina Faso
4Chronic Viral Illness Service and Division of Hematology, McGill University Health Centre, Montreal, QC, Canada
5Infectious Diseases and Immunity in Global Health Program, Research Institute, McGill University Health Centre, Montréal, QC, Canada
6Centre de recherche du Centre Hospitalier de l’Université de Montréal, Montréal, QC, Canada
7Département de microbiologie, infectiologie et immunologie, Faculté de médecine, Université de Montréal, Montréal, QC, Canada
8Département des sciences biologiques, Université de Québec à Montréal (UQAM), Montréal, QC, Canada
9Département de médecine sociale et préventive, Faculté de médecine, Université de Laval, Québec, C, Canada;
10Institut national de santé publique du Québec, Québec, QC, Canada
Caroline Gilbert
Centre de Recherche du CHU de Québec, T1-07, 2705 boulevard Laurier, Québec (QC), Canada, G1V 4G2
Phone: (418) 525-4444 ext. 46107
Fax: (418) 654-2765
E-mail: caroline.gilbert@crchudequebec.ulaval.ca
ORCID : 0000-0003-2722-1180
Wilfried Wenceslas Bazié
Centre de Recherche du CHU de Québec, T1-05, 2705 boulevard Laurier, Québec (QC), Canada, G1V 4G2
Phone: (418) 525-4444 ext. 44104
Fax: (418) 654-2765
E-mail: wilfried-wenceslas.bazie@crchudequebec.ulaval.ca
ORCID : 0000-0002-7660-9280
10.20411/pai.v6i1.384
Bazié WW, Boucher J, Vitry J, Goyer B, Routy JP, Tremblay C, Trottier S, Jenabian M, Provost P, Alary M, Gilbert C. Plasma Extracellular Vesicle Subtypes May be Useful as Potential Biomarkers of Immune Activation in People with HIV. Pathogens and Immunity. 2021;6(1):1-29. PubMed PMID. doi: 10.20411/pai.v6i1.384
Background: Extracellular vesicles (EVs) are intercellular messengers with epigenetic potential since they can shuttle microRNA (miRNA). EVs and miRNA play a role in human immunodeficiency virus (HIV) infection immunopathogenesis. Chronic immune activation and systemic inflammation during HIV infection despite effective antiretroviral therapy (ART) are associated with non-acquired immunodeficiency syndrome (AIDS) comorbidities in people living with HIV (PLWH). Analysis of plasma EVs and their miRNA content may be useful as immune activation or inflammatory biomarkers in PLWH receiving ART. In this study, we hypothesized that the number, size, and miRNA of large and small EVs could reflect immune activation associated with an elevated CD8 T-cell count or a low CD4/CD8 ratio in PLWH.
Methods: Plasma EVs subtype purified from PLWH and uninfected controls were sized using dynamic light scattering and quantified using flow cytometry and acetylcholine esterase (AChE) activity. Expression of mature miRNAs miR-92, miR-155, miR-223 was measured by quantitative reverse-transcriptase polymerase chain reaction in EVs and leucocytes.
Results: HIV infection induces increased production of small EVs in plasma. EV subtypes were differentially enriched in miR-92, miR-155, and miR-223. Positive correlations between CD8 T-cell count and large EVs abundance and small EVs AChE activity were observed. CD4/CD8 ratio was negatively correlated with small EV AChE activity, and miRNA-155 level per small EV was negatively correlated with CD8 T-cell count.
Conclusions: These findings suggest that quantifying large or small EVs and profiling miRNA content per EV might provide new functional biomarkers of immune activation and inflammation.
Keywords: biomarkers, HIV-1, extracellular vesicles, immune activation, inflammation, CD4/CD8 ratio, microRNA, miR-92, miR-155, miR-223.
Since antiretroviral therapy (ART) does not significantly reduce the generalized immune activation and inflammation during human immunodeficiency virus (HIV) infection [1, 2], it is necessary to identify factors that underlie this persistent inflammation. Reliable biomarkers of treatment effectiveness and residual immune activation and inflammation will be essential to improve patient quality of life. Potential biomarkers begin to appear in the plasma during the early stages of infection. Membranous subcellular bodies called extracellular vesicles (EVs) are secreted during communication between dendritic cells, CD4 T cells, and other immune cells that play roles in HIV infection progression [3-9]. EVs can be recovered from body fluids such as blood, plasma, serum, urine, saliva, amniotic fluid, and breast milk [10]. Since their membrane, protein, and RNA composition reflect those of the cells or tissue of their origin, EVs may indicate the disease status [11-13]. Thus, they could become biomarkers for non-invasive investigations and clinical diagnostics and prognostics for pathogenesis, immune activation, inflammatory status, and perhaps ART resistance [14].
Based on current knowledge of EV biogenesis, there are 2 main categories: exosomes or small EVs and microvesicles or large EVs [15, 16]. Exosomes are intraluminal vesicles ranging from 30 to 100 nm in diameter that are formed by the inward budding of endosomal membranes during multivesicular endosome (MVE) maturation. Exosomes are secreted when MVEs fuse with the cell surface [16, 17]. Microvesicles range from 50 to 1,000 nm in diameter and are generated by the outward budding and fission of the plasma membrane and direct release into the extracellular space [16, 18].
Previous studies have shown that EVs play critical roles in virus pathogenesis by regulating cellular functions that may inhibit or enhance viral replication [19-24]. The biological activities attributed to EVs mainly depend on their cellular origin and specific bioactive molecule content such as proteins, lipids, DNA, mRNA, microRNA (miRNA), and other non-coding RNA [25, 27]. These component levels depend mainly on the functional states of cells that secrete EVs [19]. Indeed, biological fluid EVs might precisely reflect overall immune activation or inflammatory states.
Mounting evidence suggests that EVs and short, non-coding miRNA molecules are involved in HIV infection [21, 22, 28-30]. EVs exert regulatory effects on recipient cell physiological functions via shuttled miRNA [31-34], and contact with HIV alters miRNA expression in immune cells [35, 36]. Thus, miRNA could impact HIV infection pathophysiology in at least 2 ways. First, they could bind directly to HIV-1 RNA through base-pair complementarity to interfere with HIV genomic replication [37-39] or to mRNA to block viral protein translation [29, 38, 39]. Second, miRNA could also act indirectly by targeting host cell factors called “HIV dependency factors” [40-42] or immune cell pathways that suppress or activate viral replication [43, 44]. Among the miRNA molecules believed to play some role in HIV infection, we are interested in miR-155 and miR-223, which have been implicated in immune activation and inflammation, whereas miR-92 is involved in tumorigenesis and lymphoid differentiation.
A multifunctional miRNA, miR-155 is involved in innate and adaptive immune responses such as T helper cell differentiation, particularly Th17 and regulatory T cells in people infected with HIV-1 [45, 46]. It can regulate host dependency factors involved in the trafficking and nuclear importation of pre-integration complexes such as A Disintegrin And Metalloproteinase 10 (ADAM10), Transportin-3 (TNPO3), Nucleoporin Protein 153 (Nup153), and Lens Epithelium-Derived Growth Factor/protein75 (LEDGF/p75) during HIV-1 infection [47, 48]. To date, it is the only miRNA that can distinguish elite, long-term non-progressors from naive patients, and it is upregulated in peripheral blood mononuclear cells (PBMCs) from naive HIV-1 infected patients in conjunction with pathogenesis [35]. It has also been shown to block expression of the E3 ubiquitin ligase TRIM32 by binding to the transcript’s 3’ untranslated region, which ultimately suppresses reactivation of latent HIV-1 [47, 49].
The microRNA, miR-223 is known to regulate the differentiation of several key players in the innate immune response (eg, neutrophils, monocytes, and granulocytes) and likely participates in the early stages of HIV infection and subsequent inflammation [50-52]. Its expression is positively correlated with the susceptibility of T cells, monocytes, and macrophages to infection, and this is more substantial in resting CD4 T cells (in which HIV replication is silenced) than in activated CD4 T cells. Inhibition of miR-223 or in combination with other miRNAs has been shown to enhance HIV-1 replication in resting CD4 T cells by indirectly affecting cyclin T1 levels [39, 53, 54].
The miR-92 members of the miR-17-92 cluster are distinguished as onco-miRs because of their importance in cell transformation and tumorigenesis [55]. The miR-17-92 cluster has emerged as a central integrator of gene expression events that govern T helper cell differentiation pathways. T-cell overexpression of miR-17-92 promotes proliferation and decreases the apoptosis rate, leading to lymphoproliferative disease and autoimmunity [56]. In particular, miR-92a promotes human T follicular helper cell differentiation by targeting Krüppel-like factor 2 and PTEN [57].
In silico approaches to identifying intracellular binding sites of miRNAs that directly target HIV-1 RNA have shown that miR-155 binds to vif, miR-92 to the pol region, and miR-223 to the nef region of HIV thereby contributing to repressing HIV-1 replication [37-39, 53]. Experiments have shown that miR-92 can suppress HIV-1 replication in transfected cells by 40% [38]. How miRNA contents in various EV subsets are altered under different physiological and pathological conditions must be understood both conceptually and quantitatively before their usefulness as biomarkers can be proclaimed.
Before being encapsulated in EVs for delivery to other cells, miRNAs are generated in the cell within which they will also affect post-transcriptional regulation of gene expression. PBMCs are among the major circulating targets of HIV infection, and their miRNA content profile during HIV infection has been described by several studies [29, 30]. Polymorphonuclear cells (PMNs) are a subset of immune cells and the most abundant leukocytes in human blood. They are key components of the early immune innate response to HIV infection, contribute to viremic control, and activate and modulate the quality of the adaptive immune response [58, 59]. PMNs also contribute to the chronic immune activation observed during HIV-1 infection, which is associated with the development of non-HIV/AIDS-related inflammatory conditions, even in those with well-controlled viremia under highly active ART (HAART) [59-62]. Gomez et al showed that activated neutrophils exacerbate vascular inflammation by releasing pro-inflammatory microvesicles with increased miRNA content [63]. These vesicles contain several miRNAs including regulators of inflammation (eg, miR-9, miR-150, miR-155, miR-186, miR-223, and miR-23a) with potential effects on the pathophysiological processes underpinning vascular inflammation, atherogenesis, and intestinal inflammation [63-65]. Based on these roles, PMNs also participate in EV production [66, 67] similar to other eukaryotic cells found in plasma. It is essential to understand miRNA expression in cells (PBMCs and PMNs) and the extracellular environment (EVs) during HIV infection because the value of miRNA as a biomarker could depend on where it is detected.
Despite the great interest in EVs in various pathological conditions, their isolation and nomenclature have yet to be standardized [68]. While many studies focus on exosomes, EVs are highly diverse, and it is crucial to consider differences in the secretion mechanisms and contents of their subtypes from one disease stage to the next [69]. According to our hypothesis, depending on the biogenesis mode, specific EV types contain miRNA that leads to increased CD8 T-lymphocyte counts that characterize residual immune activation in ART-treated patients. This study aimed to characterize EV subpopulations and describe miRNA content in EVs, and lymphoid, and myeloid cells. The expression levels of miR-92, miR-155, and miR-223 in large and small plasma EVs in HIV-infected and uninfected patients were investigated along with relationships between EV type, miRNA content, and clinical parameters, and between cellular and vesicular miRNA.
PLWH treated with ART with plasma viral load ≤ 50 copies/mL (n = 11) and ART-naive PLWH (n = 6) were recruited at “Unité Hospitalière de Recherche, d’Enseignement et de Soins du SIDA/VIH/hépatite (UHRESS) CHU de Québec Université Laval, CHUL.” Uninfected control participants (n = 8) were also included. The characteristics of the participating patients and healthy volunteers are presented in Table 1. Comparing immunological characteristics of PLWH, we observed that patients successfully treated with ART have lower nadir CD4 count means (± standard deviation) 259 ± 106 vs 406 ± 128 cells/µL, P = 0.0192) and longer duration of infection (18.4 ± 8.9 years vs 5 ± 4.9 years, P = 0.0031) than ART-naive patients. No significant differences were observed for CD4 T-cell count, CD8 T-cell count, or CD4/CD8 ratio. The difference in nadir CD4 count is attributable to infection duration because ART-naive patients were newly or early diagnosed (3/6), and other patients were considered long-term non-progressors (3/6).
This study received approval from the ethics review boards of Centre de recherche du CHU de Québec, Québec. All participants were anonymous volunteers and provided written, informed consent to participate in the study.
Table 1: Characteristics of participants enrolled in the study
|
ID |
Group |
Age (years) |
Sex |
Duration of HIV infection (years) |
Time on ART (years) |
CD4 nadir (cells/μL) |
HIV-1 RNA (copies/mL) |
CD4 (cells/μL) |
CD8 (cells/µL) |
CD4/CD8 ratio |
Current ART |
|
1 |
ART |
23 |
Male |
3.3 |
0.5 |
493 |
≤ 50 |
493 |
1763 |
0.28 |
ABC, 3TC, RPV |
|
2 |
ART |
60 |
Male |
27.9 |
22.1 |
187 |
≤ 50 |
481 |
635 |
0.76 |
TDF, FTC, EFV, RAL, MVC |
|
3 |
ART |
60 |
Male |
29.4 |
17.0 |
270 |
≤ 50 |
582 |
1253 |
0.45 |
TDF, FTC, NVP |
|
4 |
Control |
49 |
Male |
- |
- |
- |
- |
- |
- |
- |
- |
|
5 |
Control |
39 |
Male |
- |
- |
- |
- |
- |
- |
- |
- |
|
6 |
Naive |
29 |
Male |
8.4 |
|
341 |
11234 |
395 |
597 |
0.65 |
|
|
7 |
Control |
33 |
Male |
- |
- |
- |
- |
- |
- |
- |
- |
|
8 |
ART |
42 |
Male |
11.5 |
0.6 |
271 |
≤ 50 |
594 |
685 |
0.87 |
ABC, 3TC, RPV |
|
9 |
Control |
32 |
Female |
- |
- |
- |
- |
- |
- |
- |
- |
|
10 |
Naive |
48 |
Female |
10.1 |
- |
314 |
5608 |
470 |
1070 |
0.44 |
- |
|
11 |
Control |
56 |
Female |
- |
- |
- |
- |
- |
- |
- |
- |
|
12 |
ART |
62 |
Male |
9.5 |
8.5 |
277 |
≤ 50 |
610 |
570 |
1.07 |
TDF, FTC, EFV |
|
13 |
Control |
46 |
Female |
- |
- |
- |
- |
- |
- |
- |
- |
|
14 |
Naive |
55 |
Male |
0.2 |
- |
|
266 413 |
969 |
1352 |
0.73 |
- |
|
15 |
Control |
45 |
Female |
- |
- |
- |
- |
- |
- |
- |
- |
|
16 |
ART |
60 |
Male |
12.7 |
10.5 |
451 |
≤ 50 |
684 |
1077 |
0.64 |
ABC, 3TC, NVP |
|
17 |
Naive |
22 |
Male |
0.9 |
- |
465 |
3751 |
1433 |
3119 |
0.46 |
- |
|
18 |
Naive |
48 |
Male |
10.4 |
- |
564 |
60 |
714 |
774 |
0.91 |
- |
|
19 |
ART |
40 |
Male |
13.3 |
13.3 |
138 |
≤ 50 |
910 |
480 |
1.37 |
ABC, 3TC, LPV, RTV |
|
20 |
ART |
58 |
Male |
20.8 |
17.5 |
256 |
≤ 50 |
393 |
541 |
0.73 |
AZT, 3TC, FPV, RTV |
|
21 |
Naive |
58 |
Male |
1.3 |
- |
345 |
192300 |
432 |
902 |
0.48 |
- |
|
22 |
ART |
41 |
Male |
20.4 |
16.2 |
126 |
≤ 50 |
441 |
676 |
0.65 |
TDF, FTC, LPV, RTV |
|
23 |
ART |
60 |
Male |
25.0 |
22.4 |
76 |
64 |
491 |
757 |
0.65 |
TDF, FTC, EFV, RTV, ATV |
|
24 |
Control |
58 |
Female |
- |
- |
- |
- |
- |
- |
- |
- |
|
25 |
ART |
60 |
Male |
28.8 |
13.5 |
304 |
≤ 50 |
517 |
535 |
0.97 |
TDF, FTC, RAL |
Abbreviations: 3TC, lamivudine; ABC, abacavir; ART, antiretroviral therapy; ATV, atazanavir; AZT, zidovudine; DRV, darunavir; EFV, efavirenz; EVG, elvitegravir; FPV,fosamprenavir; FTC, emtricitabine; LPV, lopinavir; MVC, maraviroc; RAL, raltegravir; RTV, ritonavir; TDF, tenofovir disoproxil fumarate.
Peripheral blood from venipuncture was collected in EDTA-containing tubes at the CHU de Québec, QC, Canada, in 2013. PMNs and PBMCs were isolated from fresh blood samples. Cells were purified as described previously [70, 71]. Cell pellets (20 million PMNs and 5 million PBMCs) were diluted in TRIzol for miRNA analysis.
Blood obtained by venipuncture with citrate as anticoagulant was centrifuged for 10 minutes at 400
EV size was determined by hydrodynamic radius measurement using a Zetasizer Nano S (Malvern Instruments, Malvern, United Kingdom), which uses dynamic light scattering to estimate the diameter of particles in the 0.3 nm to 10 µm range. This technique is based on the fluctuation of light scattering intensity due to Brownian motion, characterized by a diffusion constant [72]. The size distribution was obtained from the distribution of diffusion constants using the Einstein-Stokes equation [72]. The size was measured in duplicate using 100 µL of EV suspension.
Acetylcholinesterase (AChE) activity was measured following a procedure described previously [13]. Briefly, EV samples (100 µL) were mixed with 1.25mM acetylthiocholine in PBS at pH 8 plus 0.1mM 5,5-dithio-bis (2-nitrobenzoic acid) in PBS at pH 7. The resulting suspension (200 µL) was held at room temperature and then warmed to 37°C for 10 minutes for optical density reading. Absorption at 450 nm was monitored for 10 minutes using a plate reader spectrophotometer (Molecular Devices with SOFTmax® Pro version 5, San Jose, CA, USA).
Thawed plasma (250 µL) was treated with proteinase K (1.25 mg/mL, Ambion™, Thermo Fisher Scientific, Waltham, MA, USA) for 10 minutes at 37°C, and EVs were recovered as described above using the method and kit described previously [13]. EV suspensions (100 µL) were diluted 3:1 in TRIzol LS (Ambion, Life Technologies, Carlsbad, CA, USA) and held at-80°C. Total RNA was extracted from EVs, PBMCs, and PMNs; mixed with 10 µL of diethylpyrocarbonate water, and quantified (1 µL) using a BioDrop-μLITE kit (Isogen Life Science, Utrecht, Netherlands). About 100 ng of RNA was treated with RNase-free DNase I (Ambion™ Life Technologies) and then reverse transcribed using a HiFlex miScript RT II kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Mature miR-92 (#MS00006594), miR-155 (#MS00031486), and miR-223 (#MS00003871) were detected by quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) using a miScript primer assay kit and miScript SYBR Green PCR kit (Qiagen). Amplification of mature miRNA as cDNA was performed in a CFX Connect real-time PCR Detection System (BIO-RAD, Hercules, CA, USA) using 40 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. Reaction specificity was confirmed using the melt curve procedure (65–95°C, 0.5°C per 5 seconds) at the end of the amplification protocol according to the manufacturer’s instructions. A standard curve was used for absolute miRNA quantification. Using synthetic hsa-miR-92, hsa-miR-155, and hsa-miR-223, an improved standard curve was obtained for miRNA concentrations ranging from 103 to 108 molecules.
A method described previously [73] for microparticle analyses was used for EV analysis by flow cytometry in a FACS Canto II Special Order Research Product cytofluorometer equipped with forward scatter coupled to a photomultiplier tube (FSC-PMT) with the “small particles option” (BD Biosciences, Franklin Lakes, NJ, USA). Purified EVs were stained with the lipophilic fluorescent carbocyanine dye DiD (DiIC18 (5) solid: 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine 4chlorobenzenesulfonate salt, Invitrogen™, Carlsbad, CA) and FITC-labeled antibodies directed against surface markers: anti-CD45, anti-CD4, and anti-CD8a (respectively products 11-9459-42, 110048-42, and 11-0089-42 from eBioscienceTM, San Diego, CA). DiD was used at 5 µL/mL of EV sample with 0.02% Pluronic F-127 (Invitrogen). After 20 minutes at 37°C, 1 portion was used to count total EVs, and the other was divided into 3 aliquots of 10µL that were each mixed with antibody and held for 15 minutes at room temperature. Next, 100 µL of 4% paraformaldehyde was added, followed by 300 µL of filtered PBS 20 minutes later and counting by flow cytometry. The gating strategy for EV identification by high-resolution flow cytometry is shown in Supplementary Figures 1 A-D.
Large EVs were separated from 250 µL of plasma, then mixed with 63 µL of ExoQuick. Total RNA was extracted from 100 µL of the small EVs and large EV fractions. Small EVs yielded 223 ng of RNA (from the 1.25X dilution due to ExoQuick), and large EVs yielded 260.6 ng (from 100 µL), which indicates that the plasma contained 2.8 ng/µL associated with small EVs and 2.6 ng/µL associated with large EVs. Based on RT-qPCR, miRNAs are expressed in copies/ng of RNA, and EV counts (obtained by cytofluorometry) are expressed per µL of plasma. Multiplying the miRNA copies/ng of RNA by the RNA per µL of plasma gives miRNA copies/µL. Dividing this result by EVs count per µL yields miRNA copies/EVs in 1 µL of plasma. By multiplying the result by a factor of 1000, we obtain the unit number of miRNA copies per vesicle in 1 ml of plasma.
Data are presented as box and whisker plots. The lines inside the boxes and the upper and lower limits of the boxes indicate the median, 75th, and 25th percentiles, respectively. The upper and lower horizontal bars indicate the maximum and minimum values, respectively. Non-parametric tests were used for comparisons. Wilcoxon matched-pairs signed-rank tests were used to compare individual paired data, and Kruskal-Wallis tests with Dunn’s multiple comparisons were used to compare group mean rankings. All correlation coefficients were calculated using Spearman rank correlation, and P values are 2-tailed. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Inc, San Diego, CA, USA). A P value ≤ of 0.05 was deemed statistically significant.
Because of the previously noted heterogeneity of EV populations [13], EVs were separated into “large” and “small” subsets to facilitate their characterization. Large EVs were pelleted from plasma after 30 minutes centrifugation at 17,000
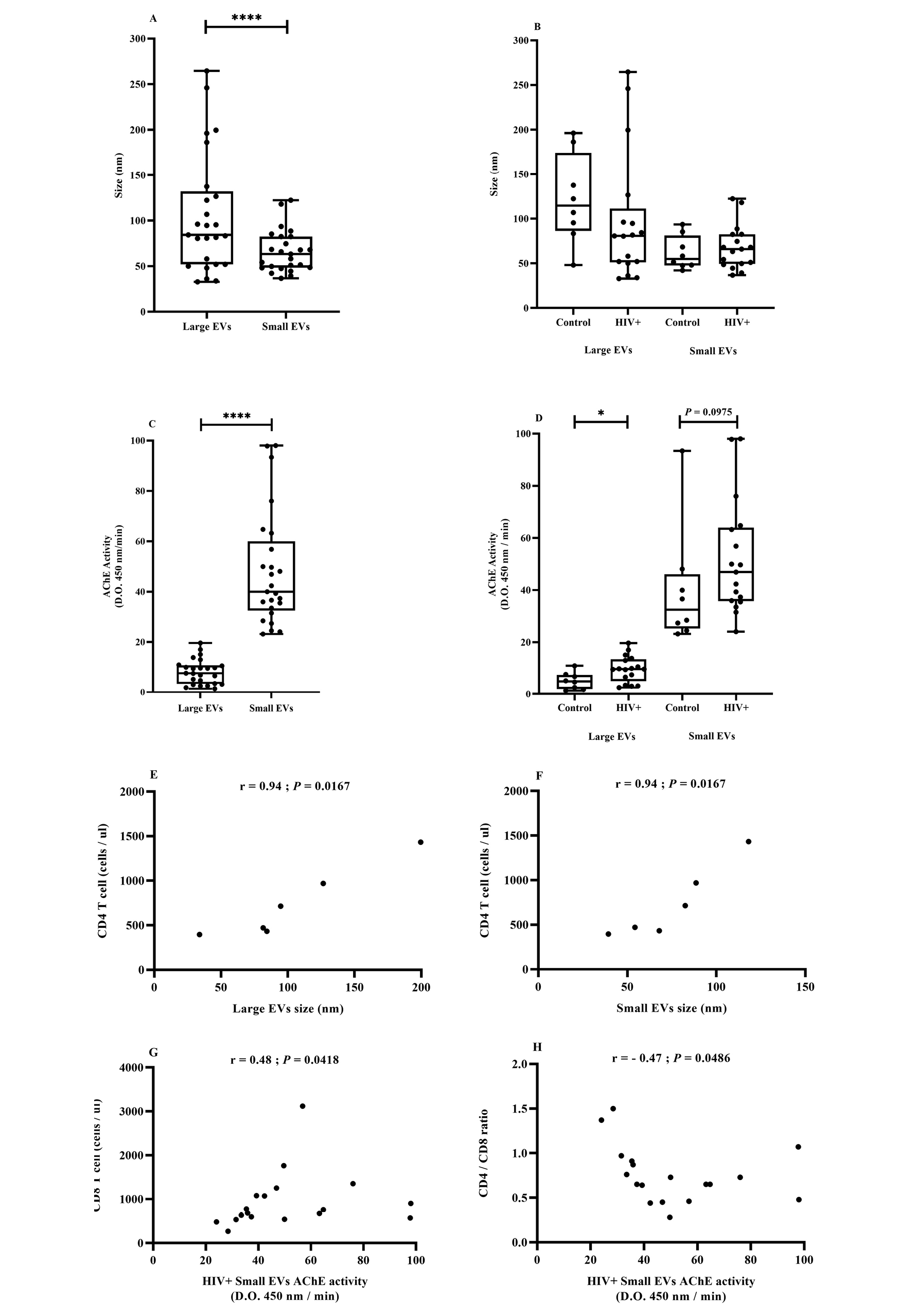
AChE activity, a generic marker of EVs of late endosome origin [74], also differentiated EV subsets. The AChE activity range was significantly greater in small EVs and did not overlap with the relatively limited range characterizing large EVs (Figure 1C). The AChE activity of large and small EVs gave means (SEM) of 8.2 (0.99) and 48.18 (4.51), respectively. The difference in AChE activity between healthy participants and HIV-infected patients was only statistically significant for the large EV fraction (Figure 1D).
The biomarker potentials of EV size and AChE activity in HIV infection were evaluated by correlation with clinical parameters. In ART-naive patients, large and small EV size was strongly and positively correlated with CD4 T-cell count (Figure 1E and F). Considering all HIV-infected participants, small EV AChE activity was positively correlated with CD8 T-cell count (Figure 1G) and inversely correlated with CD4/CD8 ratio (Figure 1H).
These correlations corroborate earlier observations [13] and reinforce the biomarker potential of plasma EVs in participants with HIV. They draw attention to the existence of EV subsets based on size and AChE activity and show that the method is useful for distinguishing both characteristics. However, AChE activity is associated with EV relative abundance and is not as good a qualitative measurement as accurate EV count and quantitative determination of cell origin.

Figure 1. EV size and AChE activity in association with HIV.
The cohort included uninfected participants (control, n = 8), patients infected with HIV (HIV+) but not treated (naive, n = 6), and patients treated successfully with antiretroviral therapy (ART, n = 11). Large and small EVs were purified as described in the Methods section. EV size distribution (Figure 1A and B) and EV AChE activity (Figure 1C and D) were evaluated respectively by dynamic light scattering and ELISA. Correlation between CD4+ T-cell count and large (Figure 1) or small (Figure 1) EV size. Correlation of CD8+ T-cell count (Figure 1G) or CD4/CD8 ratio (Figure 1H) with small EV AChE activity. Asterisks denote a significant difference (P ≤ 0.05) based on Wilcoxon matched-pairs signed-rank test.
During HIV-1 infection, several effector components of host innate and adaptive immune systems are activated to prevent the infection from gaining a foothold [75, 76]. To determine which cells shed most of the small and large EVs purified, and what impact HIV infection had on their production, EVs were counted using the lipophilic fluorescent tracer dye DiD for EV phospholipid bilayer membrane staining and antibodies directed against EV surface markers. An antibody against CD45 was used for EV leukocyte origin, and antibodies against CD4 and CD8 were used for cells bearing these receptors, particularly CD4 and CD8 T cells. We observed that the plasma concentration of small EVs was significantly higher, and this difference was observed in PLWH plasma despite their CD45+, CD4+, and CD8+ origins (Figure 2A-H).
We also measured correlations between receptor-bearing EV counts and total EV counts in the large and small vesicle categories (Figure 3). In PLWH, counts of large and small EVs bearing CD45, CD4, and CD8 were positively correlated with the total counts of large and small EVs (Figure 3 A–F). Excluding large EVs bearing CD8, these correlations were not observed in healthy participants (Figure 3A–F).
The count of cells bearing the CD8 marker was correlated with total large EV counts in ART-naive patients (Figure 3G), while the CD4/CD8 ratio had a weaker but still significant correlation with total large EV counts in the ART-treated cohort (Figure 3H). These results suggest that HIV infection induces increased production of small EVs in plasma, while associated correlations in PLWH denote more active functional participation of immune cells.
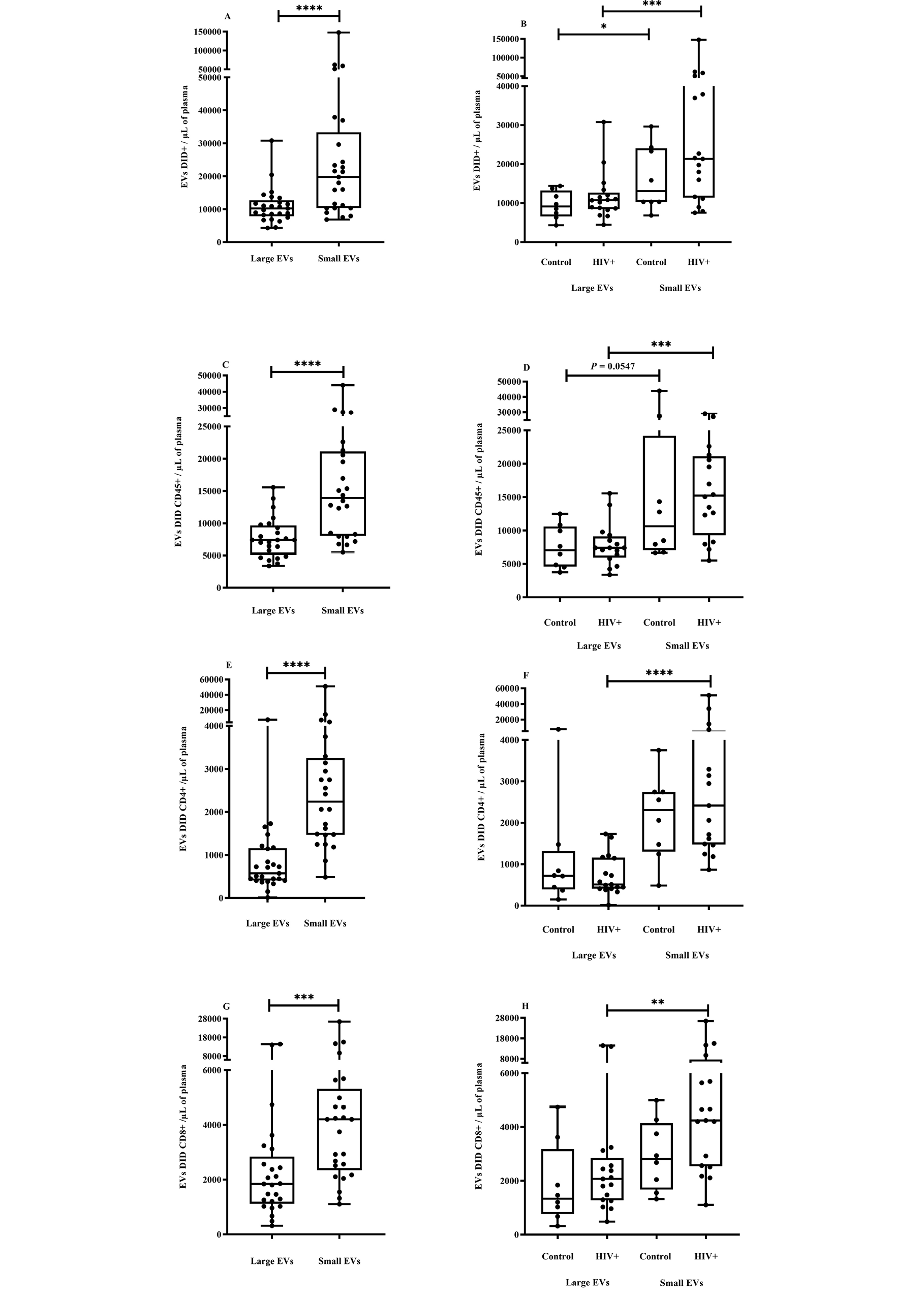
Figure 2. EV quantification and cell origin in association with HIV.
Purified plasma EVs from uninfected (Control, n = 8) and HIV+ patients (n = 17) were stained with the lipophilic fluorescent tracer dye DiD to count total vesicles (Figure 2A and B), then labeled with antibodies directed against receptors CD45 (Figure 2C and D), CD4 (Figure 2E and F), and CD8 (Figure 2 G and H) to evaluate cell origin by cytofluorometry. Asterisks denote significant difference (**P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001) based on Wilcoxon matched-pairs signed-rank test or Mann-Whitney test.
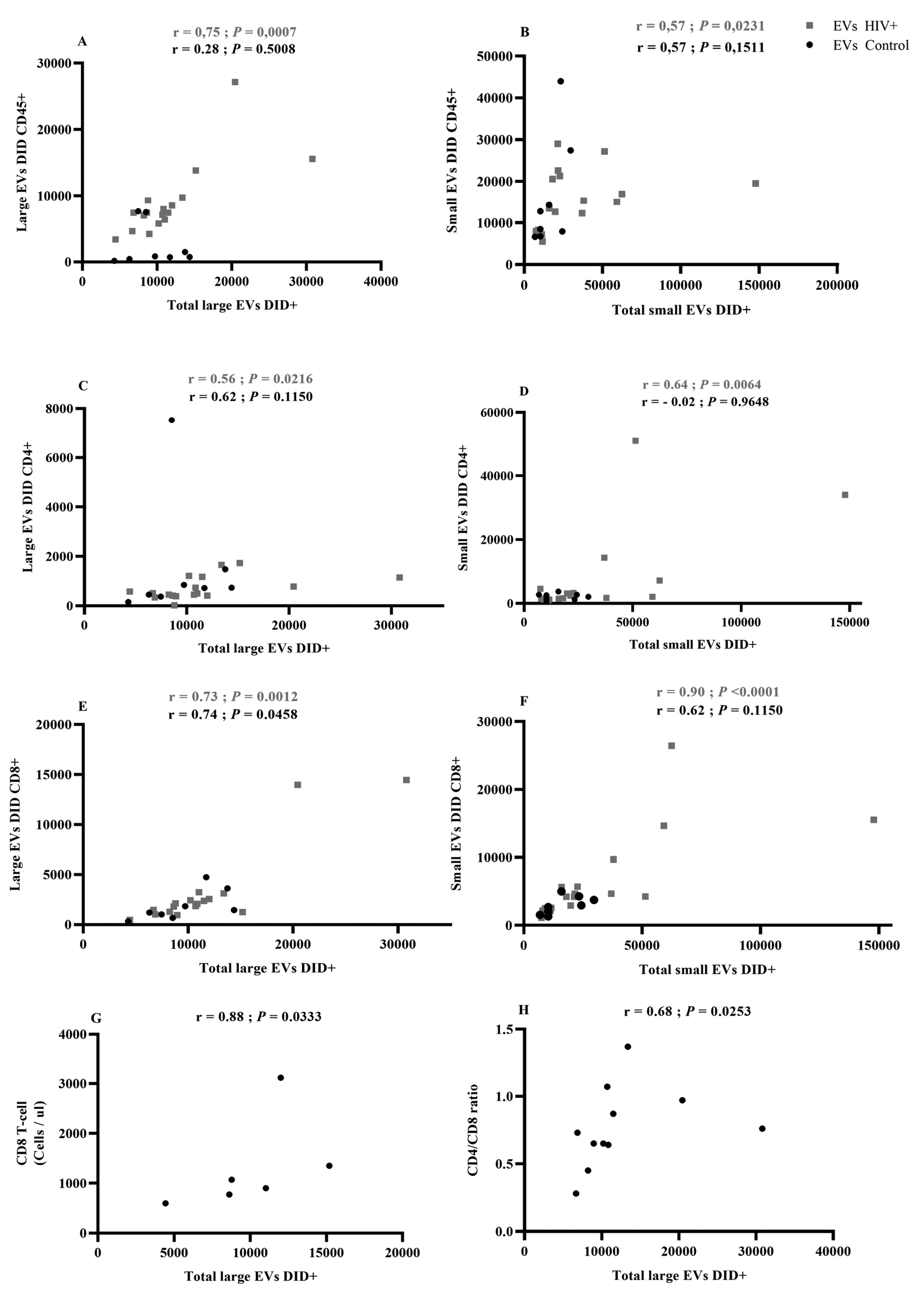
Figure 3. Correlational analysis of EV cellular origin.
A Spearman test was used to evaluate the correlation between EV total count and EVs originating from immune cells. (Figure 3 A, C, E): Large EVs bearing CD45, CD4, or CD8 correlations with total large EVs; (Figure 3 B, D, F): Small EVs bearing CD45, CD4, or CD8 correlations with total small EVs. Red indicates HIV-infected patients (HIV+, n = 17). Correlations between large EV total count and CD8 T-cell count in antiretroviral-naive patients (Figure 3G) and CD4/CD8 ratio in ART patients (Figure 3 H).

Figure 4. miRNA expression in plasma EVs in association with HIV.
Plasma samples from uninfected participants (Control, n = 8) and HIV-infected patients (HIV+, n = 17) were pretreated with proteinase K before EV purification. Total RNA was extracted from small and large EVs using TRIzol LS as per the manufacturer’s instructions and treated with DNase I. miRNA-92 (Figure 4 A and B), miR-155 (Figure Figure 4C and D) and miR-223 (Figure 4E and F) were reverse transcribed and quantified as described in the Methods section. miRNA quantity was expressed as copy number per large and small vesicle in 1 milliliter of plasma as described in the Methods section, and (Figure 4G) presents miR-155 expression as copies per vesicle. Asterisks denote significant difference (*P ≤ 0.05; **P ≤ 0.01) based on Wilcoxon matched-pairs signed-rank tests.
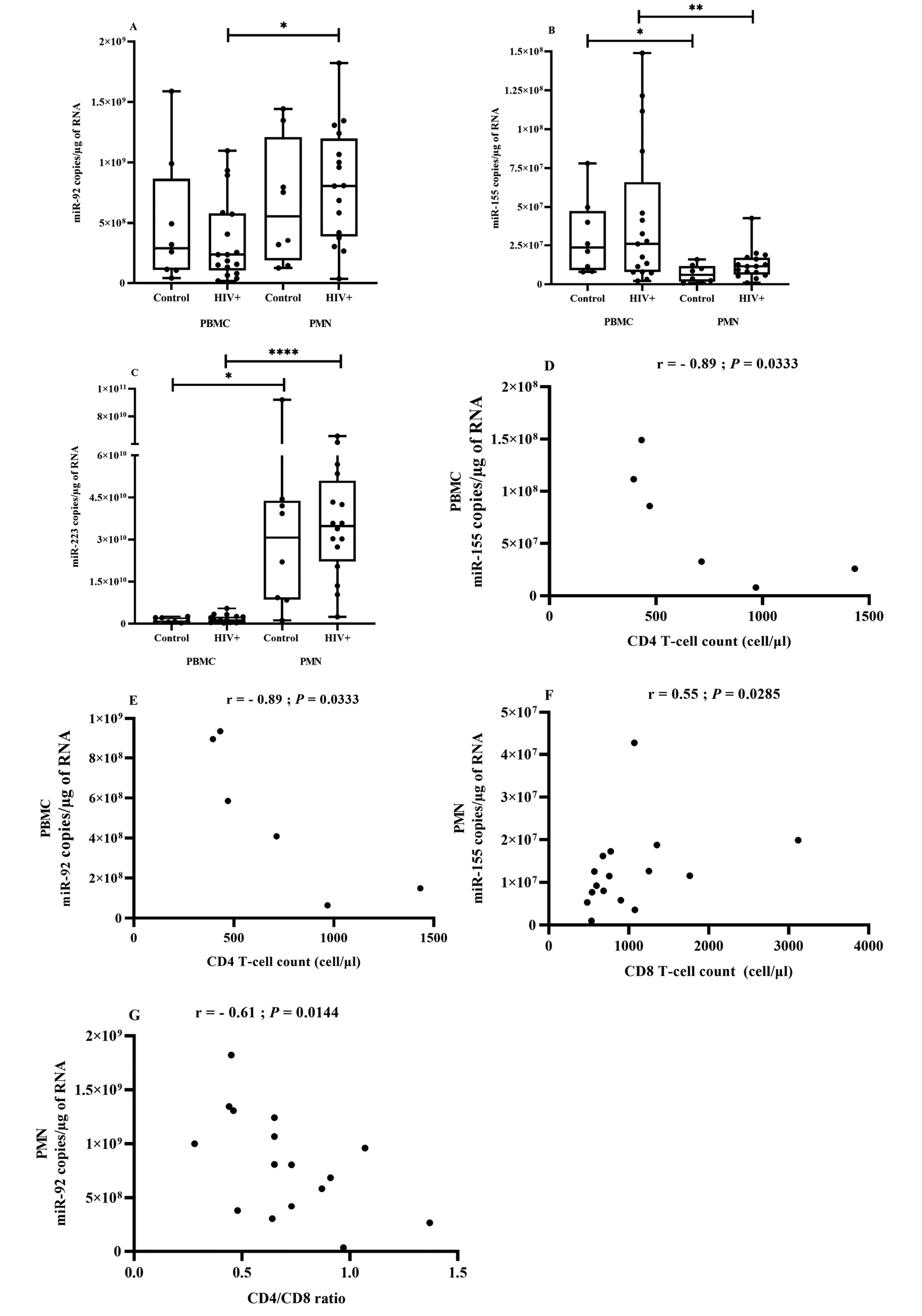
Figure 5. miRNA expression in PBMCs and PMNs in association with HIV.
Blood PBMCs and PMNs were obtained from HIV+ patients (n = 17) and control participants (uninfected, n = 8). Total RNA was isolated, and mature miR-92 (Figure 5A), miR-155 (Figure 5B), and miR-223 (Figure 5C) were quantified using RT-qPCR analysis. Comparisons are based on the Wilcoxon matched-pairs signed-rank test, and asterisks denote significant difference (*P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001). A 2-tailed Spearman test was used to evaluate the correlation between miRNA copies in PBMCs in ART-naive HIV-infected patients (n = 6) or PMNs in HIV-infected patients (n = 17) and T-cell count. (Figure 5D) miR-155 in PBMCs correlation with CD4 T-cell count, (Figure 5E) miR-92 in PBMCs with CD4 T-cell count, (Figure 5F) miR-155 in PMNs with CD8 T-cell count, (Figure 5G) miR-92 in PMNs with CD4/CD8 ratio.
Since miR-92, miR-155, and miR-223 were previously found to be more abundant in EVs from ART-naive PLWH, and miR-155 and miR-233 quantitatively correlated with EV abundance and size [13], we investigated the possibility that small and large EVs contain different amounts of these miRNA species. For this purpose, platelet-free plasma was treated with proteinase K before EV purification to eliminate plasma of free RNA bound to lipoproteins or vesicle surfaces [77]. Concerning EV miRNA content, we found that miR-92 and miR-223 were significantly more abundant in small EVs, independently of HIV status (Figure 4A, C, and E). HIV infection had no significant effect on miR-155 and miR-223 quantity in the EV subset, although the previously observed trend was preserved (Figure 4B, D, and F). The expected significant correlation between EV-borne miRNA and viral load, CD4 or CD8 T-cell count, or CD4/CD8 ratio was not found. Besides EV miRNA expression as copies per RNA microgram, we also considered the new measurement of miRNA copies per EV. We observed that miR-155 copies were more numerous in large vesicles than in small ones, and this difference was only significant in EVs from PLWH (Figure 4G and Supplementary Figure 2A and B). To evaluate the possibility of vesicular miRNA providing a new biomarker of HIV disease, we examined the relationship between EV miRNA expression level per vesicle and clinical parameters. In ART-naive PLWH, miR-155 copies per small vesicles were inversely correlated with CD8 T-cell count, although the result was not significant (Figure 4H, P = 0.0583). Copies of miR-223 per large vesicle were strongly and inversely correlated with viral load (Supplementary Figure 2C). These results suggested that some miRNAs are preferentially associated with a subset of EVs, and expression level per vesicle needs to be considered in addition to quantifying the miRNA content of EV fractions.
Despite numerous studies focused on miRNA profile changes in HIV-infected PMBCs, CD4, and CD8 T cells, PMNs have been largely neglected. We found that miR-223 and miR-92 were significantly more abundant in PMNs and miR-155 in PBMCs, but there were no significant differences between participants with HIV and controls (Figure 5A, B, and C). However, all 3 miRNA species tended to be more abundant in PBMCs from the ART-naive group.
Next, we correlated PBMC miRNA to well-known virologic and immunological markers of HIV (CD4 and CD8 T-cell counts and CD4/CD8 ratio). In ART-naive participants, miR-155 (Figure 5D) and miR-92 (Figure 5E) levels were strongly and inversely correlated with CD4 T-cell count. No correlation was found in ART patients. These results show and confirm the role of PBMCs in HIV pathology and the relevance of miRNAs as biomarkers of HIV infection.
The tendency towards increased miRNA expression in PMNs of HIV-infected patients is shown in (Figure 5A-C). No significant difference was seen between ART-naive and ART-treated patients. Spearman correlational analysis showed that increasing miR-155 in PMNs was correlated with CD8 T-cell count (Figure 5F) and increasing miR-92 was inversely correlated with CD4/CD8 ratio (Figure 5G) in association with HIV infection. The value of miRNAs as biomarkers could depend on where they are detected, and their levels in PMNs could be a new functional biomarker of inflammation.
Since the discoveries of EVs and miRNAs, evidence implicating them in human disease has mounted along with the expectation of their therapeutic and diagnostic use. In the case of miRNA, the roles of the various intracellular and extracellular forms must be elucidated before any molecule could be given serious consideration as a biomarker of HIV disease state, and the same is true for EV subsets.
Our team has shown previously that the size and AChE activity of EVs differ between PLWH and healthy participants and that these characteristics are correlated with T-cell counts and CD4/CD8 ratio [13]. To identify which EV subset is most strongly associated with these clinical parameters, we began by defining large and small size categories. In contrast with previous reports [78, 79], we found no significant difference in EV size category between uninfected participants and ART-treated or ART-naive PLWH. However, the correlation between exosome abundance (based on AChE assay) and CD8 T-cell count or CD4/CD8 ratio was corroborated in this study. AChE activity is more significant in small EVs than in large EVs and therefore has been described as a characteristic of EVs of late endosome origin (ie, exosomes) [80]. Further investigation is needed to confirm that small EVs are a reliable clinical proxy for exosomes and understand why exosomes are more abundant in HIV+ patients. Based on our results, the absence of correlation with viral load could mean that EV abundance is a marker of immune activation rather than viral replication.
Unlike AChE assays that only provide a qualitative estimation of EV relative abundance, flow cytometry provides absolute counts and quantitative detection based on fluorochrome-coupled antibodies directed against specific markers. Our results show that small EVs are more abundant than large EVs in plasma, independently of surface markers. This difference was only significant in HIV-infected patients regardless of staining (CD45, CD8, and CD4). Several studies have shown that HIV infection induces exosome release by immune cells [8, 9, 81, 82], and this could explain why CD45+, CD8+, or CD4+ EVs correlated with EV number in PLWH. Counts of EV subsets were correlated with CD8 T-cell count and CD4/CD8 ratio, and both parameters are used for clinical HIV infection management. Indeed, others have speculated that any increase in the number of circulating EVs in HIV-infected patients comes from a broad population of persistently activated immune cells [83, 84]. These results suggest that EV number could be a useful biomarker for monitoring inflammation, immune activation, and immunosenescence during HIV infection and persists despite effective ART suppression of viral replication [85-87].
Mounting evidence suggests that specific miRNAs might be clinical biomarkers of many types of diseases including HIV infection [29]. The miRNA contents of EVs are best characterized in the case of exosomes [88, 89]. Changes in the miRNA profile of HIV-infected cells are well known, and the associated EVs should reflect them. Our results show that large EVs are enriched preferentially with miR-155. We found that miR-223 is more abundant in small EVs than in large EVs. A mechanism involving RNA binding protein Y-box 1 appearing to selectively load exosomes with miRNA-223 has been previously described [89]. HIV infection might create a network in which EVs are enriched with selected miRNAs for delivery to neighboring cells. It has been estimated that a threshold concentration of at least 100 copies of miRNA must be reached to downregulate mRNA expression [90]. Our results indicate that the number of copies per vesicle is low for miR-155 compared to miR-223 and miR-92. As observed by Chevillet et al [88], this low number of miRNA copies per vesicle implied that most EVs would not contain any copies of miRNAs. These results suggest that most EVs are unlikely to be functional for miRNA transfer and raise the question of what EV type is functional for miRNA intercellular communication. However, these numbers would be sufficient for repression in a cell that captured several EVs, and EV miRNA accumulation in the recipient cell is necessary for miRNA function. Since Valadi et al [91] reported that miRNAs could be transferred between cells via exosomes, others have described antiviral functions for EVs derived from infected and uninfected cells [20, 32, 92]. Delorme-Axford et al [34] demonstrated that cultured primary human placental trophoblasts are highly resistant to infection by some viruses and, importantly, confer this resistance to nonplacental recipient cells by exosome-mediated delivery of specific miRNAs. This study highlights the functional importance of the miRNA content of EVs in an infectious context.
Cellular miRNA content during HIV infection has been suggested as a biomarker of the early/acute pathological stages [93], systemic inflammation [94], and immune activation and disease progression [13]. Many PBMC-focused studies reported cellular miRNA profile changes during HIV infection [35, 43, 54, 93, 94]. We found no difference between PLWH and control participants in terms of miR-92, miR155, and miR-223 levels associated with PBMCs and PMNs although differential regulation of miRNA in PBMCs of HIV-infected, viremic individuals, and control participants have been reported [38]. Correlations between PMN or PBMC miRNA contents and CD4 or CD8 T-cell count suggest that these molecules could provide helpful information for HIV infection management. In conclusion, this study allowed us to improve the technique of EV purification and revealed that a subset of EVs is selectively enriched in miRNA. We also demonstrated that EVs and their miRNA contents might provide clinical markers of physiological status and disease states. Although studies of EV function currently have limitations, our results show that profiling miRNA in plasma EV subsets (rather than focusing on exosomes) and miRNA expression per vesicle could provide a novel and promising approach in the context of HIV immunopathogenesis. Specifically, it could be used to assess immune activation and inflammation in treated HIV patients. As a minimally invasive tool, EV analysis could become very useful for biomedical research and the diagnosis/prognosis of immunological status or comorbidities associated with HIV infection, perhaps even leading to new immunomodulatory therapeutic targets. In the long-term, identification of biomarkers coupled with our increasing knowledge of the pathogenesis will suggest new treatment approaches specifically targeting the associated mechanism.
Supplementary materials are available at the
Conceived and designed the experiments: W.W.B. and C.G. Performed the experiments: W.W.B., C.G., J.V., J.B., and B.G. Analyzed the data: W.B. and C.G. Contributed clinical sample/reagents/materials/analysis tools: S.T., C.T., J.P.R., M.-A.J., M.A., P.P., and C.G. Wrote the paper: W.W.B. and C.G. All authors approved the final manuscript.
The authors declare that no competing interests exist. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
The authors thank Dr. Stephen Davids for assistance in editing this manuscript and constructive comments, and Dr. Martin Pelletier and Eric Boilard for access to the qPCR and EV cytofluorometry platforms, respectively. Many thanks to nurses, research assistants, Emmanuelle Rollet-Labelle, Audrey Hubert, Caroline Subra, and Alma Posvanzik for help in blood sampling and purifying PBMCs and PMN.
This study was funded through Canadian Institutes of Health Research (CIHR) grants MOP-188726; MOP-267056 (HIV/AIDS initiative) to C.G., MOP-03230 to J.P.R. and C.T. for the cohort establishment and CIHR Foundation Grant FDN-143218 to M.A. for the studentship awarded to W.W.B., as well as by the Fonds de recherche du Québec – Santé (FRQ-S) AIDS and infectious diseases network. C.T. is an FRQ-S scholar and holder of the Pfizer/Université de Montréal chair on HIV translational Research. JP.R. holds the Louis Lowenstein chair in Hematology & Oncology at McGill University. M.-A.J. holds a CIHR Canada Research Chair tier 2 in Immuno-Virology. The FRQ-S supports the Centre de recherche du CHU de Québec – Université Laval’s infrastructure.
1. Imami N, Hardy G, Burton C, Pires A, Pido-Lopez J, Moss R, Gazzard B, Gotch F. Immune responses and reconstitution in HIV-1 infected individuals: impact of anti-retroviral therapy, cytokines and therapeutic vaccination. Immunology letters. 2001;79(1-2):63-76. Epub 2001/10/12. doi: 10.1016/s0165-2478(01)00267-x. PubMed PMID: 11595291.
2. Vinikoor MJ, Cope A, Gay CL, Ferrari G, McGee KS, Kuruc JD, Lennox JL, Margolis DM, Hicks CB, Eron JJ. Antiretroviral therapy initiated during acute HIV infection fails to prevent persistent T-cell activation. Journal of acquired immune deficiency syndromes. 2013;62(5):505-8. Epub 2013/01/15. doi: 10.1097/QAI.0b013e318285cd33. PubMed PMID: 23314410; PMCID: PMC3683110.
3. Kulkarni R, Prasad A. Exosomes Derived from HIV-1 Infected DCs Mediate Viral trans-Infection via Fibronectin and Galectin-3. Scientific reports. 2017;7(1):14787. Epub 2017/11/03. doi: 10.1038/s41598-017-14817-8. PubMed PMID: 29093555; PMCID: PMC5665889.
4. Hong X, Schouest B, Xu H. Effects of exosome on the activation of CD4+ T cells in rhesus macaques: a potential application for HIV latency reactivation. Scientific reports. 2017;7(1):15611. Epub 2017/11/17. doi: 10.1038/s41598-017-15961-x. PubMed PMID: 29142313; PMCID: PMC5688118.
5. Arakelyan A, Fitzgerald W, Zicari S, Vanpouille C, Margolis L. Extracellular Vesicles Carry HIV Env and Facilitate Hiv Infection of Human Lymphoid Tissue. Scientific reports. 2017;7(1):1695. Epub 2017/05/12. doi: 10.1038/s41598-017-01739-8. PubMed PMID: 28490736; PMCID: PMC5431974.
6. Mfunyi CM, Vaillancourt M, Vitry J, Nsimba Batomene TR, Posvandzic A, Lambert AA, Gilbert C. Exosome release following activation of the dendritic cell immunoreceptor: a potential role in HIV-1 pathogenesis. Virology. 2015;484:103-12. Epub 2015/06/21. doi: 10.1016/j.virol.2015.05.013. PubMed PMID: 26092249.
7. Hubert A, Barbeau B, Subra C, Bissonnette L, Gilbert C. Role and future applications of extracellular vesicles in HIV-1 pathogenesis. Future Virology. 2015;10(4):357-70. doi: 10.2217/fvl.15.11.
8. Caroline S, Chantal B, Sophie P, Sebastien S, Caroline G. Exosomes Decrease In Vitro Infectivity of HIV-1 Preparations: Implication for CD4+T Lymphocyte Depletion In Vivo. Understanding HIV/AIDS Management and Care-Pandemic Approaches in the 21st Century2011. p. 99.
9. Subra C. Dendritic Cells Pulsed with HIV-1 Release Exosomes that Promote Apoptosis in Cd4+ T Lymphocytes. Journal of Clinical & Cellular Immunology. 2013;04(01):001. doi: 10.4172/2155-9899.S7-001.
10. Pulliam L, Sun B, Mustapic M, Chawla S, Kapogiannis D. Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer’s disease. Journal of neurovirology. 2019;25(5):702-9. Epub 2019/01/06. doi: 10.1007/s13365-018-0695-4. PubMed PMID: 30610738; PMCID: PMC7372698.
11. Lokossou AG, Toudic C, Nguyen PT, Elisseeff X, Vargas A, Rassart E, Lafond J, Leduc L, Bourgault S, Gilbert C, Scorza T, Tolosa J, Barbeau B. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells. Biology of reproduction. 2020;102(1):185-98. Epub 2019/07/19. doi: 10.1093/biolre/ioz124. PubMed PMID: 31318021.
12. Joncas FH, Lucien F, Rouleau M, Morin F, Leong HS, Pouliot F, Fradet Y, Gilbert C, Toren P. Plasma extracellular vesicles as phenotypic biomarkers in prostate cancer patients. The Prostate. 2019;79(15):1767-76. Epub 2019/09/03. doi: 10.1002/pros.23901. PubMed PMID: 31475741.
13. Hubert A, Subra C, Jenabian MA, Tremblay Labrecque PF, Tremblay C, Laffont B, Provost P, Routy JP, Gilbert C. Elevated Abundance, Size, and MicroRNA Content of Plasma Extracellular Vesicles in Viremic HIV-1+ Patients: Correlations With Known Markers of Disease Progression. Journal of acquired immune deficiency syndromes. 2015;70(3):219-27. Epub 2015/07/17. doi: 10.1097/QAI.0000000000000756. PubMed PMID: 26181817; PMCID: PMC4627170.
14. Vasconcelos MH, Caires HR, Abols A, Xavier CPR, Line A. Extracellular vesicles as a novel source of biomarkers in liquid biopsies for monitoring cancer progression and drug resistance. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2019;47:100647. Epub 2019/11/11. doi: 10.1016/j.drup.2019.100647. PubMed PMID: 31704541.
15. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual review of cell and developmental biology. 2014;30:255-89. Epub 2014/10/08. doi: 10.1146/annurev-cellbio-101512-122326. PubMed PMID: 25288114; PMCID: 25288114.
16. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of Cell Biology. 2013;200(4):373-83. Epub 2013/02/20. doi: 10.1083/jcb.201211138. PubMed PMID: 23420871; PMCID: PMC3575529.
17. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nature reviews Molecular cell biology. 2018;19(4):213-28. Epub 2018/01/18. doi: 10.1038/nrm.2017.125. PubMed PMID: 29339798.
18. Tricarico C, Clancy J, D’Souza-Schorey C. Biology and biogenesis of shed microvesicles. Small GTPases. 2017;8(4):220-32. Epub 2016/08/06. doi: 10.1080/21541248.2016.1215283. PubMed PMID: 27494381; PMCID: PMC5680703.
19. Bang C, Thum T. Exosomes: new players in cell-cell communication. The international journal of biochemistry & cell biology. 2012;44(11):2060-4. Epub 2012/08/21. doi: 10.1016/j.biocel.2012.08.007. PubMed PMID: 22903023.
20. Crenshaw BJ, Gu L, Sims B, Matthews QL. Exosome biogenesis and biological function in response to viral infections. The open virology journal. 2018;12:134–48. doi: 10.2174/1874357901812010134. PubMed PMID: 30416610; PMCID: PMC6187740.
21. Ellwanger JH, Veit TD, Chies JAB. Exosomes in HIV infection: A review and critical look. Infection, Genetics and Evolution. 2017;53:146-54. Epub 2017/05/27. doi: 10.1016/j.meegid.2017.05.021. PubMed PMID: 28546080.
22. Madison MN, Okeoma CM. Exosomes: Implications in HIV-1 Pathogenesis. Viruses. 2015;7(7):4093-118. Epub 2015/07/25. doi: 10.3390/v7072810. PubMed PMID: 26205405; PMCID: PMC4517139.
23. Meldolesi J. Extracellular vesicles, news about their role in immune cells: physiology, pathology and diseases. Clinical & Experimental Immunology. 2019;196(3):318-27. Epub 2019/02/14. doi: 10.1111/cei.13274. PubMed PMID: 30756386; PMCID: PMC6514371.
24. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nature Reviews Immunology. 2014;14(3):195-208. Epub 2014/02/26. doi: 10.1038/nri3622. PubMed PMID: 24566916; PMCID: PMC4350779.
25. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4(1):27066. Epub 2015/05/17. doi: 10.3402/jev.v4.27066. PubMed PMID: 25979354; PMCID: PMC4433489.
26. Fitzgerald W, Freeman ML, Lederman MM, Vasilieva E, Romero R, Margolis L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Scientific reports. 2018;8(1):8973. Epub 2018/06/14. doi: 10.1038/s41598-018-27190-x. PubMed PMID: 29895824; PMCID: PMC5997670.
27. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ. Reassessment of Exosome Composition. Cell. 2019;177(2):428-45 e18. Epub 2019/04/06. doi: 10.1016/j.cell.2019.02.029. PubMed PMID: 30951670; PMCID: PMC6664447.
28. Swaminathan S, Murray DD, Kelleher AD. miRNAs and HIV: unforeseen determinants of host-pathogen interaction. Immunological reviews. 2013;254(1):265-80. Epub 2013/06/19. doi: 10.1111/imr.12077. PubMed PMID: 23772625.
29. Balasubramaniam M, Pandhare J, Dash C. Are microRNAs Important Players in HIV-1 Infection? An Update. Viruses. 2018;10(3):110. Epub 2018/03/08. doi: 10.3390/v10030110. PubMed PMID: 29510515; PMCID: PMC5869503.
30. Swaminathan G, Navas-Martin S, Martin-Garcia J. MicroRNAs and HIV-1 infection: antiviral activities and beyond. Journal of molecular biology. 2014;426(6):1178-97. Epub 2013/12/29. doi: 10.1016/j.jmb.2013.12.017. PubMed PMID: 24370931.
31. Bayraktar R, Van Roosbroeck K, Calin GA. Cell-to-cell communication: microRNAs as hormones. Molecular oncology. 2017;11(12):1673-86. Epub 2017/10/13. doi: 10.1002/1878-0261.12144. PubMed PMID: 29024380; PMCID: PMC5709614.
32. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(14):6328-33. Epub 2010/03/23. doi: 10.1073/pnas.0914843107. PubMed PMID: 20304794; PMCID: PMC2851954.
33. Yoshikawa FSY, Teixeira FME, Sato MN, Oliveira L. Delivery of microRNAs by Extracellular Vesicles in Viral Infections: Could the News be Packaged? Cells. 2019;8(6):611. Epub 2019/06/21. doi: 10.3390/cells8060611. PubMed PMID: 31216738; PMCID: PMC6627707.
34. Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, Sadovsky Y, Coyne CB. Human placental trophoblasts confer viral resistance to recipient cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(29):12048-53. Epub 2013/07/03. doi: 10.1073/pnas.1304718110. PubMed PMID: 23818581; PMCID: PMC3718097.
35. Bignami F, Pilotti E, Bertoncelli L, Ronzi P, Gulli M, Marmiroli N, Magnani G, Pinti M, Lopalco L, Mussini C, Ruotolo R, Galli M, Cossarizza A, Casoli C. Stable changes in CD4+ T lymphocyte miRNA expression after exposure to HIV-1. Blood. 2012;119(26):6259-67. Epub 2012/01/31. doi: 10.1182/blood-2011-09-379503. PubMed PMID: 22286198.
36. Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2(1):81. Epub 2005/12/31. doi: 10.1186/1742-4690-2-81. PubMed PMID: 16381609; PMCID: PMC1352379.
37. Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio. 2013;4(2):e000193. Epub 2013/04/18. doi: 10.1128/mBio.00193-13. PubMed PMID: 23592263; PMCID: PMC3634607.
38. Houzet L, Klase Z, Yeung ML, Wu A, Le SY, Quinones M, Jeang KT. The extent of sequence complementarity correlates with the potency of cellular miRNA-mediated restriction of HIV-1. Nucleic acids research. 2012;40(22):11684-96. Epub 2012/10/09. doi: 10.1093/nar/gks912. PubMed PMID: 23042677; PMCID: PMC3526334.
39. Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nature medicine. 2007;13(10):1241-7. Epub 2007/10/02. doi: 10.1038/nm1639. PubMed PMID: 17906637.
40. Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathogens. 2009;5(5):e1000437. Epub 2009/05/30. doi: 10.1371/journal.ppat.1000437. PubMed PMID: 19478882; PMCID: PMC2682202.
41. Dziuba N, Ferguson MR, O’Brien WA, Sanchez A, Prussia AJ, McDonald NJ, Friedrich BM, Li G, Shaw MW, Sheng J, Hodge TW, Rubin DH, Murray JL. Identification of cellular proteins required for replication of human immunodeficiency virus type 1. AIDS research and human retroviruses. 2012;28(10):1329-39. Epub 2012/03/13. doi: 10.1089/AID.2011.0358. PubMed PMID: 22404213; PMCID: PMC3448097.
42. Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR. Host factors mediating HIV-1 replication. Virus research. 2011;161(2):101-14. Epub 2011/08/30. doi: 10.1016/j.virusres.2011.08.001. PubMed PMID: 21871504.
43. Duskova K, Nagilla P, Le HS, Iyer P, Thalamuthu A, Martinson J, Bar-Joseph Z, Buchanan W, Rinaldo C, Ayyavoo V. MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC infectious diseases. 2013;13(1):250. Epub 2013/06/01. doi: 10.1186/1471-2334-13-250. PubMed PMID: 23721325; PMCID: PMC3680326.
44. O’Connell KA, Bailey JR, Blankson JN. Elucidating the elite: mechanisms of control in HIV-1 infection. Trends in pharmacological sciences. 2009;30(12):631-7. Epub 2009/10/20. doi: 10.1016/j.tips.2009.09.005. PubMed PMID: 19837464.
45. Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunological reviews. 2013;253(1):146-57. Epub 2013/04/05. doi: 10.1111/imr.12057. PubMed PMID: 23550644.
46. Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532(1):1-12. Epub 2012/12/19. doi: 10.1016/j.gene.2012.12.009. PubMed PMID: 23246696.
47. Ruelas DS, Chan JK, Oh E, Heidersbach AJ, Hebbeler AM, Chavez L, Verdin E, Rape M, Greene WC. MicroRNA-155 Reinforces HIV Latency. Journal of Biological Chemistry. 2015;290(22):13736-48. Epub 2015/04/16. doi: 10.1074/jbc.M115.641837. PubMed PMID: 25873391; PMCID: PMC4447952.
48. Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martin S, Martin-Garcia J. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathogens. 2012;8(9):e1002937. Epub 2012/10/03. doi: 10.1371/journal.ppat.1002937. PubMed PMID: 23028330; PMCID: PMC3447756.
49. Ruelas DS, Greene WC. An integrated overview of HIV-1 latency. Cell. 2013;155(3):519-29. Epub 2013/11/19. doi: 10.1016/j.cell.2013.09.044. PubMed PMID: 24243012; PMCID: PMC4361081.
50. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125-9. doi: 10.1038/nature06607. PubMed PMID: 18278031.
51. Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. The Journal of Immunology. 2012;189(8):4175-81. doi: 10.4049/jimmunol.1201516. PubMed PMID: 22984082.
52. Haneklaus M, Gerlic M, O’Neill LA, Masters S. miR‐223: infection, inflammation and cancer. Journal of internal medicine. 2013;274(3):215-26. doi: 10.1111/joim.12099. PubMed PMID: 23772809; PMCID: PMC7166861.
53. Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113(3):671-4. Epub 2008/11/19. doi: 10.1182/blood-2008-09-175000. PubMed PMID: 19015395; PMCID: PMC2628373.
54. Chiang K, Sung TL, Rice AP. Regulation of cyclin T1 and HIV-1 Replication by microRNAs in resting CD4+ T lymphocytes. Journal of virology. 2012;86(6):3244-52. Epub 2011/12/30. doi: 10.1128/JVI.05065-11. PubMed PMID: 22205749; PMCID: PMC3302325.
55. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828-33. Epub 2005/06/10. doi: 10.1038/nature03552. PubMed PMID: 15944707; PMCID: PMC4599349.
56. Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35(2):169-81. Epub 2011/08/09. doi: 10.1016/j.immuni.2011.07.009. PubMed PMID: 21820330; PMCID: PMC3361370.
57. Serr I, Furst RW, Ott VB, Scherm MG, Nikolaev A, Gokmen F, Kalin S, Zillmer S, Bunk M, Weigmann B, Kunschke N, Loretz B, Lehr CM, Kirchner B, Haase B, Pfaffl M, Waisman A, Willis RA, Ziegler AG, Daniel C. miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(43):E6659-e68. Epub 2016/10/30. doi: 10.1073/pnas.1606646113. PubMed PMID: 27791035; PMCID: PMC5087025.
58. Casulli S, Elbim C. Interactions between human immunodeficiency virus type 1 and polymorphonuclear neutrophils. Journal of innate immunity. 2014;6(1):13-20. Epub 2013/07/23. doi: 10.1159/000353588. PubMed PMID: 23867213; PMCID: PMC6741617.
59. Yaseen MM, Abuharfeil NM, Yaseen MM, Shabsoug BM. The role of polymorphonuclear neutrophils during HIV-1 infection. Archives of virology. 2018;163(1):1-21. Epub 2017/10/06. doi: 10.1007/s00705-017-3569-9. PubMed PMID: 28980078.
60. D’Agostino C, Lichtner M, Mastroianni CM, Ceccarelli G, Iannetta M, Antonucci S, Vullo V, Massetti AP. In vivo release of alpha-defensins in plasma, neutrophils and CD8 T-lymphocytes of patients with HIV infection. Current HIV research. 2009;7(6):650-5. Epub 2009/11/26. doi: 10.2174/157016209789973600. PubMed PMID: 19929802.
61. Rajasuriar R, Khoury G, Kamarulzaman A, French MA, Cameron PU, Lewin SR. Persistent immune activation in chronic HIV infection: do any interventions work? AIDS. 2013;27(8):1199-208. Epub 2013/01/18. doi: 10.1097/QAD.0b013e32835ecb8b. PubMed PMID: 23324661; PMCID: PMC4285780.
62. Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, Hsue P, Emu B, Krone M, Lampiris H, Douek D, Martin JN, Deeks SG. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. The Journal of infectious diseases. 2008;197(1):126-33. Epub 2008/01/04. doi: 10.1086/524143. PubMed PMID: 18171295; PMCID: PMC3466592.
63. Gomez I, Ward B, Souilhol C, Recarti C, Ariaans M, Johnston J, Burnett A, Mahmoud M, Luong LA, West L, Long M, Parry S, Woods R, Hulston C, Benedikter B, Niespolo C, Bazaz R, Francis S, Kiss-Toth E, van Zandvoort M, Schober A, Hellewell P, Evans PC, Ridger V. Neutrophil microvesicles drive atherosclerosis by delivering miR-155 to atheroprone endothelium. Nature communications. 2020;11(1):214. Epub 2020/01/12. doi: 10.1038/s41467-019-14043-y. PubMed PMID: 31924781; PMCID: PMC6954269.
64. Butin-Israeli V, Bui TM, Wiesolek HL, Mascarenhas L, Lee JJ, Mehl LC, Knutson KR, Adam SA, Goldman RD, Beyder A, Wiesmuller L, Hanauer SB, Sumagin R. Neutrophil-induced genomic instability impedes resolution of inflammation and wound healing. The Journal of clinical investigation. 2019;129(2):712-26. Epub 2019/01/15. doi: 10.1172/JCI122085. PubMed PMID: 30640176; PMCID: PMC6355304.
65. Jung N, Schenten V, Bueb JL, Tolle F, Brechard S. miRNAs Regulate Cytokine Secretion Induced by Phosphorylated S100A8/A9 in Neutrophils. International journal of molecular sciences. 2019;20(22). Epub 2019/11/20. doi: 10.3390/ijms20225699. PubMed PMID: 31739406; PMCID: PMC6887701.
66. Kolonics F, Kajdacsi E, Farkas VJ, Veres DS, Khamari D, Kittel A, Merchant ML, McLeish KR, Lorincz AM, Ligeti E. Neutrophils produce proinflammatory or anti-inflammatory extracellular vesicles depending on the environmental conditions. Journal of leukocyte biology. 2020. Epub 2020/09/19. doi: 10.1002/JLB.3A0320-210R. PubMed PMID: 32946637.
67. Hong CW. Extracellular Vesicles of Neutrophils. Immune network. 2018;18(6):e43. Epub 2019/01/09. doi: 10.4110/in.2018.18.e43. PubMed PMID: 30619629; PMCID: PMC6312893.
68. Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles. 2018;7(1):1535750. Epub 2019/01/15. doi: 10.1080/20013078.2018.1535750. PubMed PMID: 30637094; PMCID: PMC6322352.
69. Valencia K, Luis-Ravelo D, Bovy N, Anton I, Martinez-Canarias S, Zandueta C, Ormazabal C, Struman I, Tabruyn S, Rebmann V, De Las Rivas J, Guruceaga E, Bandres E, Lecanda F. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Molecular oncology. 2014;8(3):689-703. Epub 2014/03/07. doi: 10.1016/j.molonc.2014.01.012. PubMed PMID: 24593875; PMCID: PMC5528646.
70. Gilbert C, Levasseur S, Desaulniers P, Dusseault AA, Thibault N, Bourgoin SG, Naccache PH. Chemotactic factor-induced recruitment and activation of Tec family kinases in human neutrophils. II. Effects of LFM-A13, a specific Btk inhibitor. Journal of Immunology. 2003;170(10):5235-43. Epub 2003/05/08. doi: 10.4049/jimmunol.170.10.5235. PubMed PMID: 12734372.
71. Ryckman C, Robichaud GA, Roy J, Cantin R, Tremblay MJ, Tessier PA. HIV-1 transcription and virus production are both accentuated by the proinflammatory myeloid-related proteins in human CD4+ T lymphocytes. Journal of Immunology. 2002;169(6):3307-13. Epub 2002/09/10. doi: 10.4049/jimmunol.169.6.3307. PubMed PMID: 12218151.
72. Hiroi T, Shibayama M. Measurement of Particle Size Distribution in Turbid Solutions by Dynamic Light Scattering Microscopy. Journal of Visualized Experiments. 2017(119):e54885. Epub 2017/01/25. doi: 10.3791/54885. PubMed PMID: 28117767; PMCID: PMC5409188.
73. Rousseau M, Belleannee C, Duchez AC, Cloutier N, Levesque T, Jacques F, Perron J, Nigrovic PA, Dieude M, Hebert MJ, Gelb MH, Boilard E. Detection and quantification of microparticles from different cellular lineages using flow cytometry. Evaluation of the impact of secreted phospholipase A2 on microparticle assessment. PLoS One. 2015;10(1):e0116812. Epub 2015/01/15. doi: 10.1371/journal.pone.0116812. PubMed PMID: 25587983; PMCID: PMC4294685.
74. Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. Journal of immunological methods. 2008;338(1-2):21-30. Epub 2008/08/05. doi: 10.1016/j.jim.2008.07.007. PubMed PMID: 18675270.
75. Altfeld M, Gale M, Jr. Innate immunity against HIV-1 infection. Nature immunology. 2015;16(6):554-62. Epub 2015/05/20. doi: 10.1038/ni.3157. PubMed PMID: 25988887.
76. McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nature Reviews Immunology. 2010;10(1):11-23. Epub 2009/12/17. doi: 10.1038/nri2674. PubMed PMID: 20010788; PMCID: PMC3119211.
77. Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Current opinion in lipidology. 2012;23(2):91-7. Epub 2012/03/16. doi: 10.1097/MOL.0b013e328350a425. PubMed PMID: 22418571; PMCID: PMC5570485.
78. Sun B, Dalvi P, Abadjian L, Tang N, Pulliam L. Blood neuron-derived exosomes as biomarkers of cognitive impairment in HIV. AIDS (London, England). 2017;31(14):F9-F17. Epub 2017/07/12. doi: 10.1097/QAD.0000000000001595. PubMed PMID: 28692534; PMCID: PMC5578870.
79. Chettimada S, Lorenz DR, Misra V, Dillon ST, Reeves RK, Manickam C, Morgello S, Kirk GD, Mehta SH, Gabuzda D. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Scientific reports. 2018;8(1):7227. Epub 2018/05/10. doi: 10.1038/s41598-018-25515-4. PubMed PMID: 29740045; PMCID: PMC5940833.
80. Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, Hill AF. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. Journal of extracellular vesicles. 2016;5(1):32945. Epub 2016/11/03. doi: 10.3402/jev.v5.32945. PubMed PMID: 27802845; PMCID: PMC5090131.
81. Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic. 2010;11(1):110-22. Epub 2009/11/17. doi: 10.1111/j.1600-0854.2009.01006.x. PubMed PMID: 19912576; PMCID: PMC2796297.
82. Benaroch P, Billard E, Gaudin R, Schindler M, Jouve M. HIV-1 assembly in macrophages. Retrovirology. 2010;7(1):29. Epub 2010/04/09. doi: 10.1186/1742-4690-7-29. PubMed PMID: 20374631; PMCID: PMC2861634.
83. Perez PS, Romaniuk MA, Duette GA, Zhao Z, Huang Y, Martin-Jaular L, Witwer KW, Thery C, Ostrowski M. Extracellular vesicles and chronic inflammation during HIV infection. Journal of extracellular vesicles. 2019;8(1):1687275. Epub 2020/01/31. doi: 10.1080/20013078.2019.1687275. PubMed PMID: 31998449; PMCID: PMC6963413.
84. Lee JH, Schierer S, Blume K, Dindorf J, Wittki S, Xiang W, Ostalecki C, Koliha N, Wild S, Schuler G, Fackler OT, Saksela K, Harrer T, Baur AS. HIV-Nef and ADAM17-Containing Plasma Extracellular Vesicles Induce and Correlate with Immune Pathogenesis in Chronic HIV Infection. EBioMedicine. 2016;6:103-13. Epub 2016/05/24. doi: 10.1016/j.ebiom.2016.03.004. PubMed PMID: 27211553; PMCID: PMC4856776.
85. Erlandson KM, Allshouse AA, Jankowski CM, Lee EJ, Rufner KM, Palmer BE, Wilson CC, MaWhinney S, Kohrt WM, Campbell TB. Association of functional impairment with inflammation and immune activation in HIV type 1-infected adults receiving effective antiretroviral therapy. The Journal of infectious diseases. 2013;208(2):249-59. Epub 2013/04/06. doi: 10.1093/infdis/jit147. PubMed PMID: 23559466; PMCID: PMC3685225.
86. Appay V, Sauce D. Immune activation and inflammation in HIV-1 infection: causes and consequences. The Journal of Pathology. 2008;214(2):231-41. Epub 2007/12/29. doi: 10.1002/path.2276. PubMed PMID: 18161758.
87. Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annual review of medicine. 2011;62:141-55. Epub 2010/11/26. doi: 10.1146/annurev-med-042909-093756. PubMed PMID: 21090961; PMCID: PMC3759035.
88. Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(41):14888-93. Epub 2014/10/01. doi: 10.1073/pnas.1408301111. PubMed PMID: 25267620; PMCID: PMC4205618.
89. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. Epub 2016/08/26. doi: 10.7554/eLife.19276. PubMed PMID: 27559612; PMCID: PMC5047747.
90. Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nature biotechnology. 2007;25(12):1457-67. Epub 2007/11/21. doi: 10.1038/nbt1372. PubMed PMID: 18026085.
91. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9(6):654-9. Epub 2007/05/09. doi: 10.1038/ncb1596. PubMed PMID: 17486113.
92. Alenquer M, Amorim MJ. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses. 2015;7(9):5066-83. Epub 2015/09/24. doi: 10.3390/v7092862. PubMed PMID: 26393640; PMCID: PMC4584306.
93. Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine. 2019;43:307-16. Epub 2019/04/22. doi: 10.1016/j.ebiom.2019.04.023. PubMed PMID: 31005516; PMCID: PMC6557912.
94. Ballegaard V, Ralfkiaer U, Pedersen KK, Hove M, Koplev S, Braendstrup P, Ryder LP, Madsen HO, Gerstoft J, Gronbaek K, Nielsen SD. MicroRNA-210, MicroRNA-331, and MicroRNA-7 Are Differentially Regulated in Treated HIV-1-Infected Individuals and Are Associated With Markers of Systemic Inflammation. Journal of Acquired Immune Deficiency Syndromes. 2017;74(4):e104-e13. Epub 2016/10/18. doi: 10.1097/QAI.0000000000001191. PubMed PMID: 27749601.
Submitted May 14, 2020 | Accepted November 2, 2020 | Published January 14, 2021
Copyright © 2021 Pathogens and Immunity. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International License.